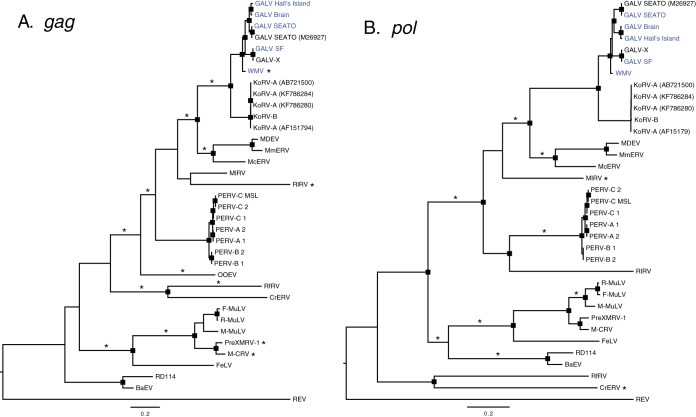
FIG 6.
Maximum-likelihood phylogenetic trees of gammaretroviruses inferred using complete pol (A) and gag (B) nucleotide sequences. Node robustness was assessed with 500 rapid bootstrap pseudoreplicates. The rectangles on the nodes indicate a bootstrap support of >80%. The GALV strain sequences generated in this study are highlighted in blue. Branches with significant (P < 0.05) evidence of episodic diversifying selection, as indicated by the BSREL method, are marked with an asterisk. GenBank accession codes are shown in brackets. The scale bar indicates 0.2 nucleotide substitutions per site. The trees are midpoint rooted for purposes of clarity. All abbreviations can be found in Table 2.