FIG 1.
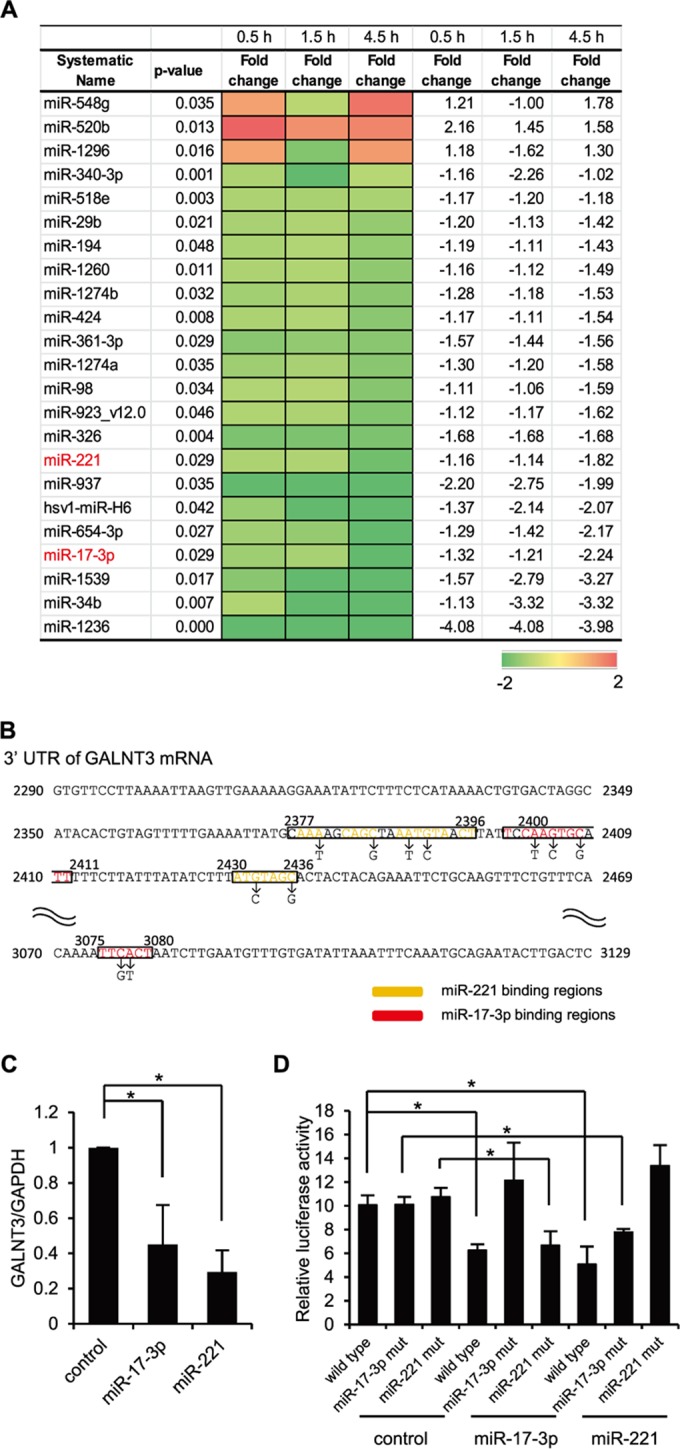
Downregulation of miR17-3p and miR-221 during the early stage of IAV infection. (A) A heat map comparing the fold changes of microRNAs with significantly higher (red) or lower (green) expression in A549 cells infected with PR8 virus at an MOI of 1.0. The P values for the fold changes are indicated. (B) Sequences of predicted miR-17-3p and miR-221 binding sites in the 3′ UTR of GALNT3 mRNA. The black letters below the predicted binding sites (boxed) indicate the nucleic acid substitutions in the mutant plasmids used for the luciferase assay. (C) The reduction of GALNT3 mRNA expression by miR-17-3p and miR-221 mimic oligonucleotides. HEK293T cells were transfected with oligonucleotides mimicking miR-17-3p and miR-221, and the expression of GALNT3 mRNA was detected by real-time PCR. The relative expression levels of GALNT3 and GAPDH are shown (control, 1.0). Control indicates scrambled control miRNA. (D) miR-17-3p and miR-221 target the 3′ UTR of GALNT3 mRNA. Plasmids containing point mutations in the putative miR-17-3p and miR-221 binding sites or miR-17-3p mut and miR-221 mut were transfected into HEK293T cells after the introduction of the mimic oligonucleotide. The luciferase activity in the transfected cells was monitored 24 h after transfection. The y axis indicates the luciferase activity of the pRL-tk reporter relative to that of the pGL3 control plasmid. All values are expressed as the means (n > 3) ± standard errors of the means (SEM). Statistical significance was analyzed by Student's t test. *, P < 0.05.
