Figure.
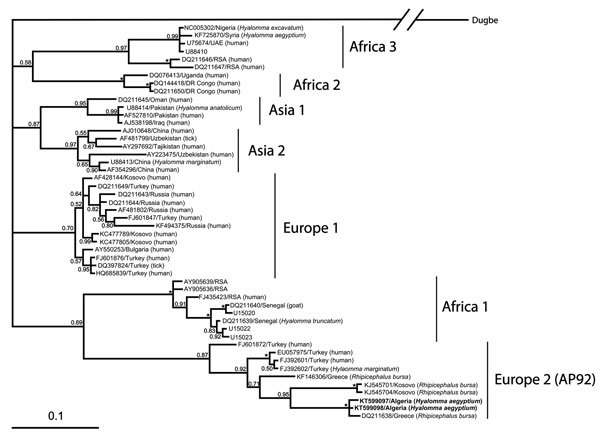
Phylogenetic analysis of Crimean-Congo hemorrhagic fever virus small RNA segment sequences, performed by using Bayesian inference in MrBayes version 3.1.2. (http://mrbayes.csit.fsu.edu/) under a general time-reversible plus gamma distribution plus invariable site model with 107 generations setup. Bootstrap values (>50%) are shown at nodes. Asterick (*) indicates 1.00 bootstrap value. Scale bar represents the estimated number of substitutions per site. Individual sequences are named with GenBank accession number/country of origin and the host, if available, in parentheses. Boldface indicates sequences of virus isolated from ticks collected from 12 Testudo graeca tortoises in Algeria, 2009–2010.
