The RNA-binding domains of Arabidopsis SRSF1 splicing factors are required for nucleocytoplasmic dynamics and protein-protein interactions.
Abstract
Ser/Arg-rich (SR) proteins are essential nucleus-localized splicing factors. Our prior studies showed that Arabidopsis (Arabidopsis thaliana) RSZ22, a homolog of the human SRSF7 SR factor, exits the nucleus through two pathways, either dependent or independent on the XPO1 receptor. Here, we examined the expression profiles and shuttling dynamics of the Arabidopsis SRSF1 subfamily (SR30, SR34, SR34a, and SR34b) under control of their endogenous promoter in Arabidopsis and in transient expression assay. Due to its rapid nucleocytoplasmic shuttling and high expression level in transient assay, we analyzed the multiple determinants that regulate the localization and shuttling dynamics of SR34. By site-directed mutagenesis of SR34 RNA-binding sequences and Arg/Ser-rich (RS) domain, we further show that functional RRM1 or RRM2 are dispensable for the exclusive protein nuclear localization and speckle-like distribution. However, mutations of both RRMs induced aggregation of the protein whereas mutation in the RS domain decreased the stability of the protein and suppressed its nuclear accumulation. Furthermore, the RNA-binding motif mutants are defective for their export through the XPO1 (CRM1/Exportin-1) receptor pathway, but retain nucleocytoplasmic mobility. We performed a yeast two hybrid screen with SR34 as bait and discovered SR45 as a new interactor. SR45 is an unusual SR splicing factor bearing two RS domains. These interactions were confirmed in planta by FLIM-FRET and BiFC and the roles of SR34 domains in protein-protein interactions were further studied. Altogether, our report extends our understanding of shuttling dynamics of Arabidopsis SR splicing factors.
Ser/Arg-rich (SR) protein is the collective name given to a family of highly conserved splicing factors in Eukaryotes that regulate constitutive and alternative precursor mRNA splicing. SR proteins contain at least one RNA recognition motif (RRM) and an Arg/Ser-rich (RS) C-terminal domain (Manley and Krainer, 2010; Califice et al., 2012). The RRM appears to determine RNA-binding specificity, while the RS domain is involved in protein-protein and protein-RNA interactions (Shen et al., 2004). In human, twelve SR proteins have been described based on a set of formal criteria (Manley and Krainer, 2010). SR proteins have a modular organization: some SR proteins contain two RRMs while others contain a Zn-knuckle, which contributes to RNA binding. The activity of SR proteins is regulated by posttranslational modifications, such as Ser phosphorylation/dephosphorylation and Arg methylation. At steady-state, SR proteins accumulate in subnuclear speckles, which correspond to storage, assembly, and/or modification compartments for splicing factors. Several human SR proteins shuttle between the nucleus and the cytoplasm, and this dynamic shuttling is linked to their postsplicing activities in mRNA export, stability, and translation (Long and Caceres, 2009). The multiple roles and mechanisms of action of mammalian SR proteins have been extensively studied (for review, see Long and Caceres, 2009; Zhong et al., 2009; Kornblihtt et al., 2013; Änkö, 2014).
The number of genes encoding SR proteins is higher in plants compared with metazoan. Plant genomes contain SR proteins homologous to the animal prototypes SRSF1/SRSF2/SRSF7, as well as plant-specific ones (Barta et al., 2010; Califice et al., 2012). Arabidopsis SR splicing factors localize into nuclear irregular dynamic domains similar to speckles, with no, only partial or complete colocalization (Tillemans et al., 2005; Lorković et al., 2008; Reddy et al., 2012). The functions of plant SR factors in postsplicing events remain unknown, though a nucleocytoplasmic shuttling activity has been described for RSZ22, a prototypic member of the SRSF7 subgroup (1 RRM, 1 Zn-knuckle) of Arabidopsis (Arabidopsis thaliana) SR protein family (Tillemans et al., 2006; Rausin et al., 2010).
The nucleocytoplasmic transport of RNA and proteins occurs through nuclear pore complexes (NPCs), which require importin and exportin receptors (karyopherins or Kap) for trafficking of molecules larger than 40–90 kD. Kap often binds to cargo molecules that carry either nuclear localization signals (NLS) for nuclear import or nuclear export signals (NES) for nuclear export (Boruc et al., 2012). The best-known import pathway is mediated by the importin-α/β Kap that binds to NLS. Kap-β2 (or Transportin-SR, TRN-SR) was shown to function as the nuclear import receptor for human SRSF1 and SRSF2, and several Arabidopsis SR proteins (Yun et al., 2003; Xu et al., 2011). The human TRN-SR has recently been shown to embrace both the RRM and RS domains of SRSF1 for nuclear import (Maertens et al., 2014).
XPO1 (Exportin-1, also named CRM1 in yeast [Saccharomyces cerevisiae]) is a well-characterized mammalian nuclear export receptor which recognizes Leu-rich NES (φ-X2-3-φ-X2-3-φ-X-φ, where φ is L, V, I, F, or M and X is any amino acid) on proteins implicated in snRNA and rRNA export (Natalizio and Wente, 2013). XPO1/CRM1 was also shown to mediate the export of unspliced (or partially spliced) viral mRNAs and of a small subset of mRNAs. XPO1 recruitment to mRNA is mediated by single adaptor proteins including Leu-rich pentatricopeptide repeat proteins (LRPPRC) and HuR (Natalizio and Wente, 2013). Apart from this, the bulk of mRNA is exported by the nonkaryopherin heterodimer Nxf1-Nxt1 (TAP-p15) in metazoans (Mex67-Mtr2 in yeast). The shuttling SR proteins are known to promote messenger ribonucleoprotein (mRNP) export through NPCs when dephosphorylated by interacting with export factor Nxf1 (Huang et al., 2003). Several human SR proteins are also part of the exon junction complex (EJC) deposited upstream of exon-exon junctions after splicing, consistent with a role of SR proteins in mRNP export and nonsense mediated RNA decay (Singh et al., 2012). The RS domain is necessary but not sufficient for the cytoplasmic export of shuttling SR proteins (Cáceres et al., 1997).
We previously identified RSZ22 as a shuttling splicing factor whose nuclear export is at least partly controlled by the XPO1-dependent export pathway (Tillemans et al., 2006; Rausin et al., 2010). Mutating conserved residues within the RNA-binding motifs of this specific SR protein highlighted the in vivo dependence of RNA binding for proper subcellular dynamics (Rausin et al., 2010). However, the role of the different protein domains in directing the cellular dynamics may vary among SR proteins, and the role of the RS domain of RSZ22 had not been investigated. It is also unknown whether XPO1-dependent nuclear export also includes other Arabidopsis SR proteins. A more global understanding of the molecular mechanisms underlying the nucleocytoplasmic transport of plant SR factors therefore required further investigation.
Here, we functionally characterized the four Arabidopsis SR proteins of the SRSF1 subfamily (orthologs of mammalian SRSF1) that contain two conserved RRM domains (Califice et al., 2012). We studied the expression profiles of SR30, SR34, SR34a, and SR34b, and attempted to investigate their shuttling activity. Among these SR proteins, SR30 showed a less active nuclear export rate, and SR34b protein was not detectable in any expression assay. Because of its stability and rapid shuttling, we further focused on the SR34 protein by generating a series of mutant versions of the RRMs and RS domains. We established the overall requirement of these protein domains to retain nucleocytoplasmic shuttling activity. Yeast two-hybrid (Y2H) assays also revealed strong interactions between SRSF1 subfamily members (SR30, SR34, and SR34a) and SR45, an atypical SR protein (two RS domains). We also investigated the importance of SR34 domains in protein-protein interactions. Collectively, our findings provide a more detailed mechanistic understanding of the role of the structural determinants regulating SR proteins dynamics, and insights into protein domain function in in vivo interactions.
RESULTS
Expression Analysis of Arabidopsis SR Factors of the SRSF1 Subfamily
The Arabidopsis SRSF1 subfamily contains four members, SR30, SR34, SR34a, and SR34b. To date, few studies have characterized their expression profiles and protein dynamics (Lopato et al., 1999; Lopato et al., 2002; Fang et al., 2004). We have undertaken a global analysis of their expression as a prerequisite to dynamic studies. Quantitative RT-PCR showed that the four genes were expressed in all Arabidopsis vegetative and floral organs examined with slightly different expression levels for SR30, SR34, and SR34a (Fig. 1A). SR34b was weakly expressed and the SR34b protein was not detected in plant cells (nor in yeast, see below), supporting the hypothesis that SR34b is a pseudogene (Kalyna and Barta, 2004). The spatial expression patterns of SR30, SR34, and SR34a were investigated using GUS reporter constructs (PSR30:GUS, PSR34:GUS, and PSR34a:GUS) and GFP translational fusions (PSR30:SR30-GFP, PSR34:SR34-GFP, and PSR34a:SR34a-GFP). Several independent promoter-reporter T3 lines were generated for each SR gene that all showed identical staining/fluorescence patterns.
Figure 1.
Expression profiles of Arabidopsis SRSF1 subfamily members. A, Quantitative RT-PCR analyses of SR30, SR34, SR34a, and SR34b gene expression in Arabidopsis vegetative and floral organs. Data (mean ± SEM) are normalized transcript levels relative to At1g58050 (see “Materials and Methods”). Detection of GUS activity (blue staining) directed by the SR30 promoter (B), SR34 promoter (C), SR34a promoter (D) in root, leaf, and floral tissues. Data were analyzed by one-way ANOVA followed by Tukey multiple comparison tests (P < 0.05). Statistically significant differences between means of all genes within one tissue are indicated by different superscripted letters, whereas differences between means for one gene between tissues are indicated by different superscripted numbers.
The GUS expression profiles corroborated the quantitative RT-PCR results. In young seedlings, expression of the GUS reporter for SR30 was observed in cotyledons and root tip. A faint staining could be observed in root epidermal cells. During vegetative growth, GUS activity was visualized in young leaves and primary and lateral roots (root tips and stele with a stronger signal at lateral root initiation). As leaves expand, GUS was constrained to vascular tissues (primary, secondary, and tertiary veins) and hydathodes (Fig. 1B). GUS staining was not observed in any other cell types of fully differentiated leaves, including branched trichomes and stomata. During floral development, PSR30:GUS expression looked uniform in unopened floral buds. From stage ∼ 10–12, GUS staining was more intense in the receptacle at the base of the flowers and within the style. In buds where only sepals had opened, GUS was detected in the sepal vascular tissues, in style and in anther filaments. Once the flower was fully open, staining was apparent in the upper part of the anther filament and pollen grain, which increased further in mature pollen. In developing siliques, GUS was visualized in funiculus (Fig. 1B).
SR34 and SR34a showed similar expression profiles during vegetative growth in shoots and roots. PSR34:GUS and PSR34a:GUS activity was observed in young seedlings in both cotyledon and primary roots where staining was high at the level of the root tip including meristem, and in the stele. Young leaf primordia were very intensely stained. In young and expanding leaves, GUS staining was typically seen in the veins, hydathodes, and in cells at the base of trichomes and trichomes. In older leaves, GUS expression resolves to the margins and particularly in vascular tissue and hydathodes.
In inflorescences, P34:GUS activity was observed in inflorescence stems and in unopened floral buds. From stage ∼ 10–12, GUS staining was visible within the sepals and the style. In buds where only sepals had opened, GUS was detected in the sepal vascular tissues, in style and in anther filaments. Once the flower was fully open, staining was apparent in the upper part of the anther filament and pollen grain, which increased further in mature pollen. In developing siliques, GUS was visualized in funiculus (Fig. 1C).
PSR34a:GUS showed a faint staining in unopened floral buds up to stage 6 to 7. Later, GUS activity was observed in sepals, petals, and became high in the style. As stamen matured, expression was seen in filaments and developing pollen. At later floral stages, GUS activity was observed in upper part of filaments, in stigmatic papillae, in pollen and germinating pollen, and in ovule funiculi (Fig. 1D).
Heterozygous and homozygous translational fusion (Px:SRx-GFP) lines were indistinguishable from wild-type plants, indicating that the expression of either transgene did not alter plant development. The localization of the translational fusions were in agreement with the GUS expression profiles described above (Supplemental Fig. S1). SRSF1 subfamily members were observed exclusively in nuclei of specific cell types, however, with different quantitative abundance despite relatively high and similar mRNA expression levels determined by quantitative real-time RT-PCR (see above). GFP translational fusion proteins showed different expression levels with much weaker expression for SR30-GFP than SR34-GFP and SR34a-GFP. Indeed, in all analyzed tissues, fluorescence of SR30-GFP was very weak and sometimes hardly detectable above the background.
Nucleocytoplasmic Shuttling of the SRSF1 Subfamily Members in Arabidopsis Transgenics
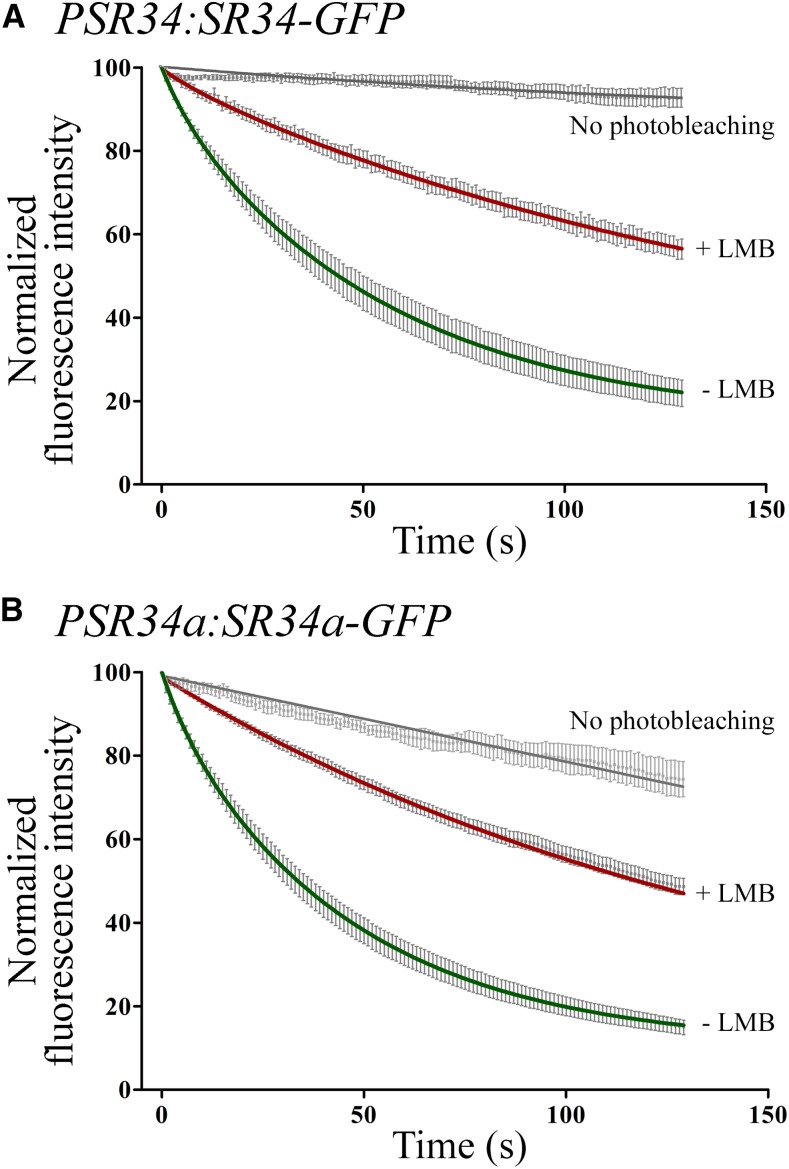
Next, we analyzed the dynamic shuttling of SRSF1 members in transgenic Arabidopsis plants using a fluorescence loss in photobleaching (FLIP)-shuttling assay, which measures the exchange of GFP between nucleus and cytoplasm (Tillemans et al., 2006; Rausin et al., 2010). As mentioned above, the observed fluorescence emission in the PSR30:SR30-GFP homozygote plants was very weak and the fluorescence of unbleached control cells decreased very strongly during time-lapse experiments. Hence, SR30 export shuttling kinetics could not be established with accuracy. By contrast, nucleocytoplasmic shuttling of SR34 and SR34a was apparent from FLIP curves in root cells of PSR34:SR34-GFP and SR34a:SR34a-GFP plants (Fig. 2). Our FLIP data were best fitted with two-phase exponential decay curves, suggesting the presence of two distinct nuclear populations for each SR splicing factor with different half-lives (i.e. slow half-lives of ∼ 38 s and ∼ 32 s and fast half-lives of ∼ 5 s and ∼ 4.78 s for SR34 and SR34a, respectively, with a R2 of ∼ 0.7).
Figure 2.
Nucleocytoplasmic shuttling of SR34 and SR34a in Arabidopsis transgenics. FLIP-shuttling of SR34-GFP (A) and SR34a-GFP (B) was monitored in the absence (−LMB) and upon LMB (+LMB) treatment in root cells. One hundred percent fluorescence indicates prebleach fluorescence intensity. As a control, cells were repeatedly scanned under no photobleaching conditions and fluorescence was quantified. Half-time of fluorescence decay for –LMB and +LMB curves were as follows (in seconds): ∼ 38 and ∼ 80 (A), ∼ 32 and ∼ 78 (B), respectively. Values are means ± SEM for at least 16 nuclei. The curves show a significant inhibitory effect of LMB on shuttling (P < 0.0001).
The interaction between XPO1 and NES can be inhibited by leptomycin B (LMB), inducing nuclear accumulation of the shuttling proteins (Tillemans et al., 2006; Rausin et al., 2010). Root cells of transgenic GFP reporter lines treated with LMB showed that reducing XPO1 export pathway resulted in decreased shuttling kinetics of both SR34-GFP and SR34a-GFP proteins (Fig. 2). One exponential decay curves fitted the experimental data (SR34 and SR34a half-lives of ∼ 80 s and ∼ 78 s, respectively). These results demonstrate that SR34 and SR34a splicing factors shuttle rapidly between the nucleus and cytoplasm and suggest that they exhibit an intrinsic XPO1-dependent shuttling activity as previously revealed for RSZ22 (Rausin et al., 2010).
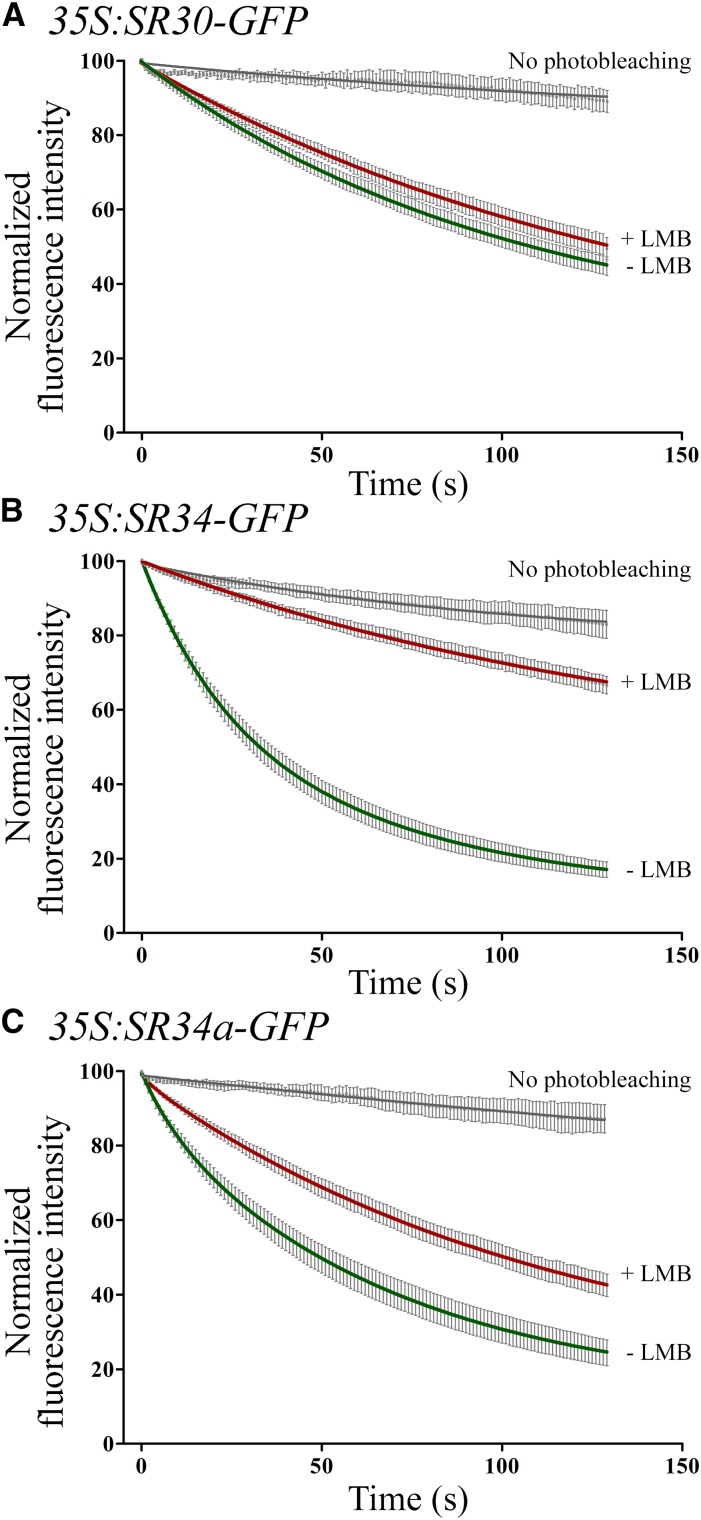
As previously shown, a transient expression assay can also accurately evaluate SR protein shuttling activity (Tillemans et al., 2006; Rausin et al., 2010). Thus, we further examined nucleocytoplasmic shuttling of wild type SRSF1-like proteins in tobacco leaf cells transiently expressing SR genes under the control of the constitutive CaMV35S promoter. This allowed sufficient SR30 protein expression for shuttling analysis. The SR30 protein showed a low rate of nuclear export with a single exponential fluorescence decay (half-life of ∼ 71 s). When cells were treated with LMB, the mobility of SR30 was weakly affected (Fig. 3A; half-life of ∼ 90 s). In the same experimental set-up, the SR34b protein could not be detected suggesting its instability. Finally, lifetime decays of SR34 and SR34a were fit to two-exponential decay curves, as above. SR34 export was significantly reduced upon LMB treatment whereas the shuttling of SR34a was also blocked by LMB albeit to a lesser extent (Fig. 3, B and C). These results suggest that SR34 and SR34a shuttle more rapidly between the nucleus and cytoplasm than SR30 in transient assays and that SR34 (and SR34a) shuttling is at least partially XPO1-dependent in transient expression assays.
Figure 3.
Nucleocytoplasmic shuttling of SRSF1 subfamily members in transient expression assays in tobacco leaf cells. FLIP-shuttling of SR30-GFP (A), SR34-GFP (B) and SR34a-GFP (C) was monitored in the absence (−LMB) and upon LMB (+LMB) treatment in leaf epidermal cells. One hundred percent fluorescence indicates prebleach fluorescence intensity. As a control, cells were repeatedly scanned under no-photobleaching conditions and fluorescence was quantified. Values are means ± SEM for at least 13 nuclei. The FLIP curves show a significant inhibitory effect of LMB on shuttling for SR34 and SR34a (P < 0.0001).
SR34 Domains Mediating XPO1-Dependent Nucleocytoplasmic Shuttling
A more comprehensive understanding of the nucleocytoplasmic shuttling of plant SR proteins in vivo requires a thorough analysis of the roles of RNA-binding and RS domains in this process. As for wild-type proteins, transient expression assay can accurately assess whether and how SR protein domains contribute to shuttling activity (Tillemans et al., 2006; Rausin et al., 2010).
We intended to monitor the dynamics of a series of SR protein mutants. To achieve this goal, SR34-GFP was selected because of its rapid nucleocytoplasmic shuttling, strong fluorescence, and apparent XPO1-dependent export. As all proteins of the SRSF1 subfamily, SR34 has two types of RRM: an N-terminal canonical RRM and a central pseudo-RRM containing the invariant SWQDLKD motif (Califice et al., 2012). The RS domain is extended by a PSK motif (Pro/Ser/Lys-rich) of unknown function (Reddy, 2004). These specific domains of SR34 are schematically represented in Supplemental Figure S2.
RRMs are all characterized by a β1α1β2β3α2β4 fold but the mode of binding to RNA nucleotides involves different broadly conserved residues in canonical RRM domain (RNP1 and RNP2 motifs) and pseudo-RRM domain (SWQDLKD motif; Cléry et al., 2013). Structural data of plant SR proteins (RRM and pseudo-RRM domains) are currently not available. To support the design of our mutagenesis experiments, both SR34 RRM domains were modeled and a clear similar overall topological arrangement can be observed as shown in Supplemental Figures S3 and S4. In SR34 RRM1 domain, Y10 and F49 form stacking pi interactions with RNA nucleotides. The aromatic character of these residues is broadly conserved in RRM domains and were shown to be indispensable for RNA-binding capacity (Cléry et al., 2008; Daubner et al., 2013). Other SR34 residues potentially interacting with RNA (D37, K39, Y47, and E78) are broadly conserved in plants and animal splicing factors (Califice et al., 2012), suggesting the binding to a common RNA fragment. It is worth noting that the loop between β2 and β3 strands is made of four prolines and one Arg (PPRPP), and is highly characteristic of, and specific to, plant splicing factors (Califice et al., 2012).
The mode of binding to RNA of pseudo-RRM domains was recently elucidated by solving the NMR structure of human SRSF1 RRM2 bound to a UGAAGGAC RNA fragment (Cléry et al., 2013). Pseudo-RRMs lack the aromatic residues involved in RNA binding of canonical RRM, and their interaction with RNA is mediated by a highly conserved heptapeptide SWQDLKD, which specifically binds to a GGA motif. Whether SR34 RRM2, with its conserved heptapeptide, binds to a GGA motif as well is not known.
We mutagenized highly conserved aromatic residues mediating RNA binding in the RNP1 and RNP2 motifs of the canonical N-ter RRM domain. The rnp1 mutant consisted of two point mutations of the aromatic Tyr and Phe residues (Y47A and F49A), and the rrm1 mutant combined those mutations to a Y10A substitution in rnp2. The rrm2 mutant consisted of two point mutations of the Trp and Phe residues (W133A and F148A). The mutations within each RRM were combined to generate the rrm1/rrm2 mutant protein. It has been previously shown with human proteins that such aromatic residue mutations severely impair the RNA-binding capacity of the RRM1 and RRM2 (Cáceres and Krainer, 1993; Heinrichs and Baker, 1997; Gama-Carvalho et al., 2001; Tintaru et al., 2007). To assess the role of the RS domain in the functional dynamics of SR34, all Ser and/or Arg residues within a 197-279 fragment were substituted to Thr and/or Gly, respectively (Cazalla et al., 2002), generating the GS, RT, and GT mutants. PSK alterations consisted in point mutations of all Ser into Thr in this C-ter motif (Supplemental Fig. S2). These SR34 mutant proteins fused to GFP were transiently expressed in Arabidopsis and tobacco leaf cells, and similar results were obtained.
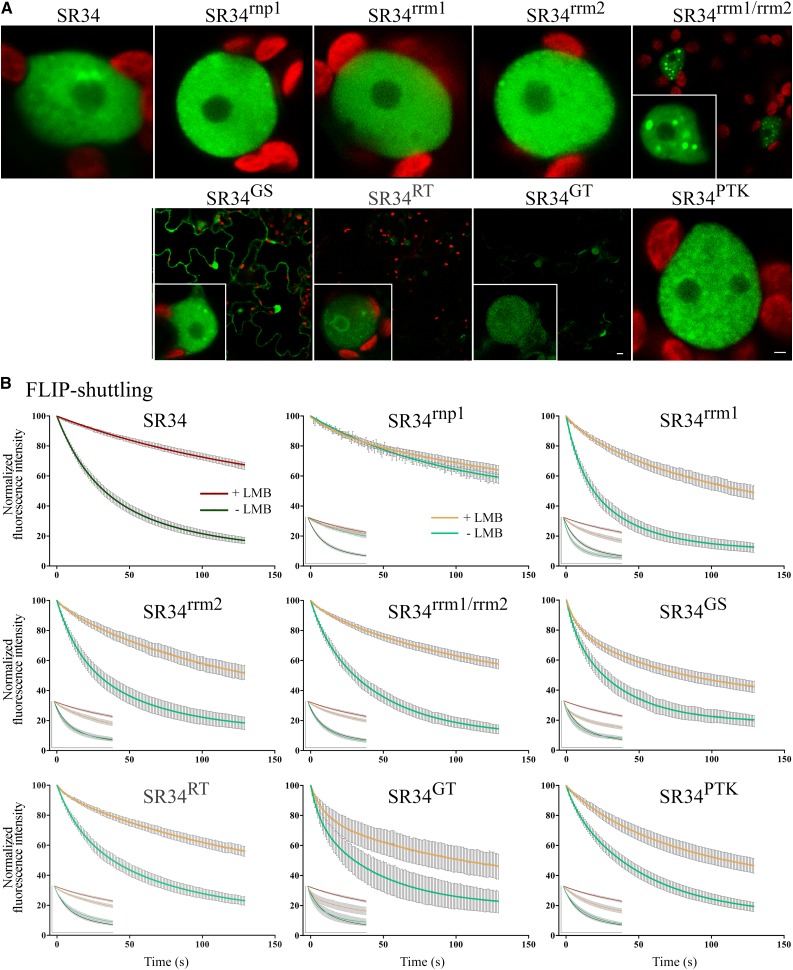
Upon transient expression in tobacco leaf cells, all RRM mutant proteins were exclusively nuclear often with a less-pronounced speckled localization, and the mutation of both RRMs led to an important aggregation of SR34rrm1/rrm2 in large nuclear subcompartments (Fig. 4A). The Arg substitutions within the RS domain resulted in nuclear speckled and cytoplasmic localization of SR34GS. In contrast, SR34RT was less stable than SR34 and exclusively nuclear with often abnormal aggregation at the perinucleolar compartment. SR34GT displayed additive effect of Ser and Arg substitutions and was both nuclear and cytoplasmic but barely detectable (Fig. 4A). SR34PTK was exclusively nuclear but displayed a high number of small speckled-like structures (Fig. 4A).
Figure 4.
Dynamic localization of SR34 and mutant derivatives in transient expression assays in tobacco leaf cells. A, Selected images of nuclear fluorescence distribution of GFP-tagged SR34 (far left) and mutant proteins. Bars = 1 µm. B, Nucleocytoplasmic shuttling of SR34 (top left) and mutant proteins. FLIP-shuttling was monitored in the absence (−LMB) and upon LMB (+LMB) treatment. Insets show the overlay of wild-type and mutant curves. Values are means ± SEM for at least 20 nuclei. The differences observed in the FLIP curves between untreated and LMB treated cells are statistically different for SR34 and SR34rrm1/rrm2 (P < 0.0001), SR34RT (P < 0.001), SR34rrm1, and SR34rrm2 (P < 0.01).
Next, FLIP-shuttling studies were performed on tobacco leaf fragments to assess the nuclear export of SR34 mutant variants. These experiments showed similar rapid rates of nuclear export of SR34-GFP and all mutant proteins except SR34rnp1. Intriguingly, the mutation of the sole RNP1 motif strongly inhibited the export of the protein (Fig. 4B). Similar data were obtained with SR34rnp1 transiently expressed in Arabidopsis leaf cells, albeit with an export rate slightly higher due to a more effective photobleaching of the cytoplasm during FLIP assay in Arabidopsis leaf cells (Supplemental Fig. S5). All mutant variants (except SR34rnp1) retained higher nucleocytoplasmic export than SR34 upon LMB treatment. The localization of the SR34 variants was not affected by LMB treatment (Fig. 4B). As expected, the export rate of SR34rrm1/rrm2 and SR34RT was slightly lower than the other mutants due to protein aggregation, reducing their overall mobility (Fig. 4A).
Protein-Protein Interactions of the SRSF1 Subfamily Members
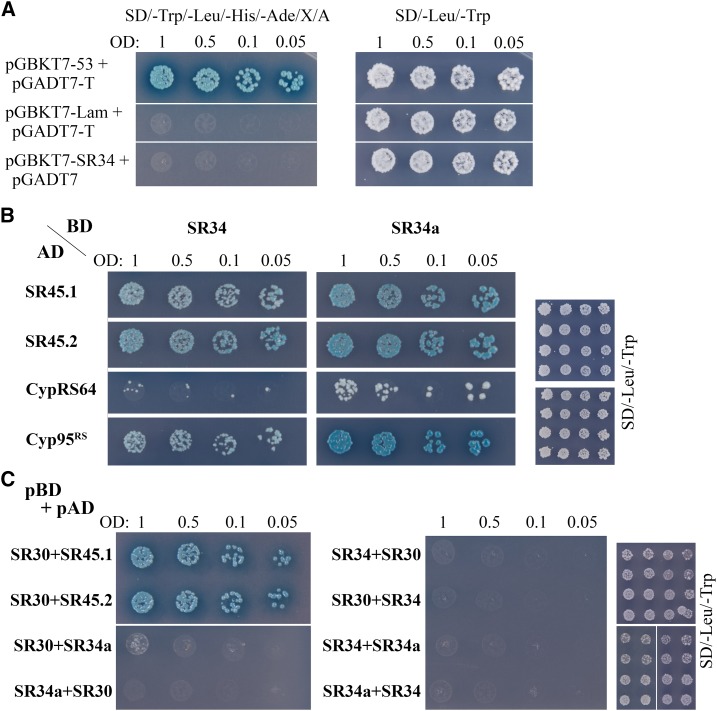
In order to understand the molecular mechanisms involved in SR34 protein-protein interactions, we conducted a yeast two-hybrid screen using SR34 as bait. The screen identified different proteins already known to interact with SR34, i.e. CypRS64 (At3g63400), the RS domain of Cyp95 (At4g32420) and SRPK4 (At3g53030; Lorkovic et al., 2004; de la Fuente van Bentem et al., 2006; Fig. 5). Interestingly, more than 95% of the identified clones contained the full-length open reading frames of the two isoforms of SR45, SR45.1 and SR45.2, suggesting a strong interaction between SR34 and SR45 (Fig. 5). In a targeted approach, we were also able to detect direct interactions between SR34a and all these proteins (Fig. 5B). SR30 was only tested for interaction with SR45 (SR45.1 and SR45.2), and we did not observe interaction in yeast between SR30, SR34, and SR34a (Fig. 5C).
Figure 5.
Arabidopsis SRSF1 subfamily members interactions detected by yeast two-hybrid analysis. From the mated culture, dilutions to an OD600 of 1, 0.5, 0.1, and 0.05 were spotted on synthetic dropout (sd)/-Trp/-Leu/-His/-Ade/X-α-Gal/AurA agar plates. Positive interactions were confirmed by growth and blue staining. Yeast cultures on SD/-Leu /-Trp control plates confirm the presence of both plasmids (right). A, Mated diploids between Y2HGold harboring pGBKT7-53 and Y187 containing pGADT7-T were used as positive control. pGBKT7-Lam and pGADT7-T or pGBKT7-SR34 and pGADT7-AD, in either case, were used as negative controls. B, Mating between Y2HGold containing pGBKT7-SR34 or SR34a and Y187 harboring pGADT7- SR45.1, SR45.2, CypRS64 or Cyp95RS. SR34 and SR34a both interact with SR45.1, SR45.2, CypRS64 or Cyp95rs in yeast cells. C, Mating test between different baits (pGBKT7, pBD) SR34, SR34a or SR30 and preys (pGADT7, pAD) SR34, SR34a, SR30, SR45.1 or SR45.2. SR30 interacts with both SR45 isoforms but not with SR34 and SR34a. SR34 and SR34a do not interact.
In order to determine which SR34 domains are involved in the identified protein complex formation, we tested whether and how SR34 mutations described above affected protein-protein interactions. The strength of interaction could be assessed by measuring the ability of yeast diploids to grow in auxotrophic conditions. Mutations of either the RRM2 or the PSK motif did not affect the interactions between the mutant derivatives and both SR45 and SRPK4 (Supplemental Fig. S6). In contrast, SR34rnp1 displayed weaker interaction with both SR45 and SRPK4. SR34rrm1 and SR34rrm1/rrm2 completely lost their interaction with SR45 but had stronger interaction with SRPK4 (Supplemental Fig. S6). The substitution of the Arg residues into Gly resulted in weaker interaction with SR45 and in loss of detectable interaction with SRPK4. The substitution of the Ser residues into Thr resulted in weaker interaction with SR45 but in stronger interaction with SRPK4 (Supplemental Fig. S6).
The interaction between SR34 and SR45 was further assessed in living plant cells by two complementary approaches, namely bimolecular fluorescence complementation (BiFC) and fluorescence resonance energy transfer (FRET) based on fluorescence lifetime imaging (FLIM). First, BiFC assays were performed in tobacco leaf cells that transiently coexpressed SR34-Yellow Fluorescent Protein N-terminal fragment (YFPN) and SR45-YFP C-terminal fragment (YFPC), or reciprocally SR34-YFPC and SR45-YFPN. As shown in Supplemental Figure S5, YFP fluorescence appeared in the nuclei with a speckled-like distribution, suggesting SR34/SR45 interaction in planta. As controls, no fluorescence was detected in cells transfected with SR34-YFPN and YFPC, or SR34-YFPC and YFPN (Supplemental Fig. S7A).
A robust way of analyzing FRET in living cells is the measurement of the excited state lifetime of the donor fluorophore by FLIM. FLIM was used to measure FRET, and hence interaction, between SR34 and SR45 tagged with GFP (donor) and mCherry (acceptor), respectively. First, we measured the average fluorescence lifetime of untagged GFP and of SR34-GFP upon transient expression in tobacco cells. The fluorescence lifetime of unquenched GFP was well fitted with the use of a single exponential model for free GFP and SR34-GFP with a χ2 close to one, yielding τGFP = 2.49 ± 0.01 ns (mean ± sd for n = 6) and τSR34-GFP = 2.43 ± 0.01 ns (n = 6), respectively (Supplemental Fig. S7B). In cells coexpressing the bound complexes, SR34-GFP and SR45-mCherry, the average fluorescence lifetime of SR34-GFP significantly decreased to 2.05 ± 0.05 ns (n = 6) and the fluorescence decay exhibited double exponential model in which the long lifetime was fixed to the lifetime of the donor alone SR34-GFP (2.43 ns) and the short lifetime corresponded to the FRET lifetime (1.21 ± 0.1 ns; Supplemental Figure S7B). We also measured donor lifetime in cells coexpressing SR34rrm1-GFP/SR45-mCherry and SR34rrm2-GFP/SR45-mCherry. Cells coexpressing SR34rrm1-GFP and SR45mCherry exhibited donor fluorescence decay well-fitted with a single exponential model (τSR34rrm1-GFP = 2.42 ± 0.04 ns for n = 5) indicating the absence of FRET and, hence, no interaction. The average fluorescence lifetime of SR34rrm2-GFP in cells coexpressing SR45-mCherry was reduced compared with cells only expressing SR34-GFP or SR34rrm1-GFP (τSR34rrm2-GFP = 2.05 ± 0.06 ns for n = 5), and two exponential components yielded a satisfactory fitting of the fluorescence lifetime decay (Supplemental Fig. S7B). As FRET standard and positive control, the sensitivity of FLIM in vivo in transient expression assay was tested using cells expressing the tandem GFP-mCherry, and we found an average lifetime of 2.05 ± 0.03 ns with a long lifetime 2.4 ns and a short lifetime of 1.02 ns (Supplemental Fig. S7B). Together, these data confirm the yeast two-hybrid studies and provide compelling evidence of SR34/SR45 interaction and the direct involvement of the SR34 RRM1 in this interaction.
Nuclear Export of SR45
Our study supports the direct association of SR45 with SRSF1-subfamily members in vivo, but whether SR45 is a shuttling protein has not been characterized. Therefore, we investigated the shuttling activity of SR45 in transient assay using cytoplasmic FLIP as described above. In SR45-GFP expressing cells, the nuclear fluorescence was efficiently decreased with time, indicating that SR45-GFP is exported from the nucleus (Supplemental Fig. S8). The export rate of SR45-GFP could be fitted to two exponential decay curve (slow half-life of ∼ 74 s and fast half-life of ∼ 16 s). Upon LMB treatment, the kinetics of nuclear export was strongly reduced with a one exponential decay curve fitting the experimental data (half-life of ∼ 102 s). Our data suggest the involvement of XPO1 in nuclear export of SR45.
DISCUSSION
In eukaryotes, macromolecular complexes larger than 40 to 60 kD are actively transported through NPCs, and therefore, gene-expression regulation requires a regulated and dynamic nucleocytoplasmic transport of molecules (Boruc et al., 2012; Field et al., 2014). The nascent premRNA transcripts recruit shuttling RNA-binding proteins, including SR proteins. A subset of the human SR protein family members are nucleocytoplasmic shuttling proteins that have roles in a wide-range of postsplicing processes, such as mRNA export, stability, and mRNA translation. The mechanisms involved in SR protein export in plants and the possible roles of their domains in this process are poorly understood. We previously showed that RSZ22 shuttles between nucleoplasm and cytoplasm in a XPO1 (CRM1)-dependent manner (Tillemans et al., 2006; Rausin et al., 2010). We also provided evidence that RSZ22 RNA-binding domains play a role in regulating nuclear export activities through the XPO1 pathway. XPO1 is the most conserved exportin across eukaryotes (Serpeloni et al., 2011), and in Arabidopsis, two XPO1 isoforms (XPO1a and XPO1b) have been identified (Merkle, 2003). Whether XPO1 regulates the nuclear export of (sub)populations of mRNAs in plants remains unknown (see below).
In this report, we examined the dynamic nucleocytoplasmic shuttling of members of the SRSF1 subfamily (two RRMs) of Arabidopsis SR proteins. Our data support that among the Arabidopsis SRSF1-like proteins, SR30 and SR34a are less actively exported out of the nucleus in transient-expression assays, and this export appears to be almost totally XPO1-independent for SR30. However, as discussed previously (Rausin et al., 2010), cytoplasmic FLIP in transient assays may not fully reflect the exact dynamics of SR proteins. Indeed, we cannot totally rule out that distinct SR proteins are less effective in processing (pre)mRNAs in transient expression assays. The expression of SR30 under the control of its endogenous promoter was quantitatively too weak to establish accurate SR30 shuttling kinetics in transgenic plants. To assess shuttling kinetics of less-actively exported (and/or less expressed) splicing factors would require to adapt the FLIP approach or to set up other sensitive real-time shuttling assays in plant cells. In transient expression assay, FLIP-shuttling is performed on leaf cells containing large vacuoles and relatively low cytoplasmic content. Repeatedly bleaching larger, or the entire, cytoplasmic area might lead to the monitoring of nuclear fluorescence over time. Alternatively, the fluorescence could only be monitored before and after the FLIP time-lapse to evaluate putative loss of fluorescence, but it would not provide FLIP kinetic curve. Moreover, single-molecule fluorescence (SMD) methods and fluorescence correlation spectroscopy have proven to be valuable to provide insights on transport of cargo molecules in permeabilized and intact cells (Cardarelli et al., 2012; Goryaynov et al., 2012), However, such new methodology development was beyond the scope of the present report. In contrast, SR34 exits the nucleus by active transport and partially accumulates in the nucleus upon XPO1 inhibition in transient ectopic- and stable-tissue-specific expression assays. Importantly, the integrity of SR34 RRMs is necessary for XPO1-dependent nuclear export, since mutations of conserved residues in either RRM1 or RRM2 reduced the inhibitory effect of LMB. The SR34 nuclear export mediated by XPO1 thus appears RNA-binding-dependent, similar to our previous results on RSZ22 (Rausin et al., 2010). Mutations of each individual SR34 RNA-binding domain (either RRM1 or RRM2) did not fully inhibit the nuclear export of mutant proteins, even upon LMB treatment, suggesting the passive diffusion of SR34rrm1 and SR34rrm2 through the NPCs.
Mutations of both RRMs appear to profoundly perturb subnuclear localization of the mutant SR34rrm1/rrm2, inducing its aggregation. Intriguingly, the mutation of the aromatic residues (Y and F) of the sole RNP1 motif of SR34 induced a significant and substantial nuclear retention of the mutant protein. How the RNP1 is implicated in nuclear export is unclear. The present data suggest that this precise motif is important for protein-protein interactions since mutations of RNP1 weaken the interaction between SR34rnp1 and both SR45 and SRPK4 in yeast and plant cells. We could speculate that the nuclear export of SR34 is tightly tied to its ability to interact with other splicing factors, including SR45 (this study), but it would not explain the absence of passive diffusion of the SR34rnp1 through NPCs (visible upon LMB inhibition). SR34rrm1 was not retained in the nucleus, yet RRM1 mutation did not impair all protein-protein interactions (SR34rrm1/SRPK4). It is worth mentioning that the residues just upstream of the RNP1 motif, forming the loop between the β2 and β3 strands, differ between animal (RRGGP) and plant splicing factors (PPRPP; Supplemental Fig. S3). The conformation of this loop, which borders the nucleotide binding zone, is probably different between animal and plant splicing factors, and we do not know how this difference influences binding affinity to nuclear components. These data call for structural studies of plant SR protein domains allowing to establish relationships between specific amino acid substitutions and structural alterations.
The RS domain of several mammalian SR proteins (i.e. SRSF1, SRSF2, and SRSF3) has been shown to function as an NLS. The cellular (and subnuclear) localization of SR proteins is regulated by the reversible phosphorylation of Ser residues within the RS domain by at least two protein kinase families, SRPKs (SR-specific kinases) and CLKs (Cdc2-like kinases). The cytoplasmic SRPK1 phosphorylates the N-terminal stretch of RS repeats (called RS1) in the RS domain of SRSF1. The C-terminal RS domain (RS2) contains a Ser/Prorich region that can be phosphorylated by nuclear CDK1 leading to hyperphosphorylation of SRSF1 (Aubol et al., 2013). The RS domains of plant SR proteins are also targets for phosphorylation, and SRPK4 was found to phosphorylate three sites of RS31 in vitro that were determined to be phosphorylated in vivo (de la Fuente van Bentem et al., 2006; Barta et al., 2008). The SR proteins SR34 and SRZ21 have been shown to be phosphorylated in vitro by MAPK family members (Feilner et al., 2005). The position of phosphorylation sites within SR34a were located in the short C-terminal PSK motif (de la Fuente van Bentem et al., 2006). Interestingly, our findings demonstrate a distinctive role of Ser residues of the RS domain and of the C-terminal PSK motif of SR34. We show that the substitution of Ser into Thr within the RS domain of SR34 induced a severe instability of the mutant protein. Interestingly, SR34RT could still strongly interact with SRPK4 but our preliminary results suggest that SR34RT was not phosphorylated in vivo. However, despite its instability, SR34RT was still nuclear in contrast to the human SRSF1, which requires phosphorylation of the RS1 domain for nuclear import by Transportin-SR (Lai et al., 2001; Maertens et al., 2014). Structural studies of the phosphorylated RRM2-RS1 domain of SRSF1 associated to Transportin-3 (Tnpo3) revealed an interaction between Ser phosphates and Arg residues of Tnpo3 (Maertens et al., 2014). However, not all SR proteins are imported into the nucleus in a phosphorylation-dependent manner (Yun et al., 2003). Whether plant SR proteins are transported actively as animal SR proteins remains unknown. In Arabidopsis, two TRN-SR of the Tpno3 family have been identified (Tamura and Hara-Nishimura, 2014). MOS14 (modifier of snc1-1, 14;) was identified as one of these, and the loss-of-function of the MOS14 isoform results in altered splicing patterns of SNC1 and RPS4, two resistance genes implicated in plant immunity (Xu et al., 2011). Y2H assays showed that MOS14 interacts with SR proteins (including SR34) via its C terminus (Xu et al., 2011). We showed that Arg mutation of RS domain abolished nuclear import of SR34GS consistent with the observation that the RS domain of SR proteins serves as a nuclear localization signal (Tillemans et al., 2006; Reddy et al., 2012). We observed the inability of SR34GS to interact with SRPK4. Therefore, SRPK(4)-mediated phosphorylation might play an important role in regulating nuclear import of SR34 and other plant SR proteins. Interestingly, the PSK motif of SR34 appears to be important for the nucleocytoplasmic shuttling dynamics of the protein, but substitution of Ser residues did not affect the stability of SR34PTK and its nuclear localization, nor its ability to interact with SR45 and SRPK4 in yeast. Thus the PSK appears to possess unique features independently of the RS domain.
During recent years, it has become increasingly apparent that the mechanisms controlling mRNA nucleocytoplasmic export is highly complex in mammals and yeast (Opisthokonta), involving a variety of factors (Natalizio and Wente, 2013). The mRNA export depends on the recruitment of the conserved TRanscription and EXport (TREX) complex which is formed by the association of many factors including the RNA helicase Uap56 (Sub2 in yeast), the RNA-binding adaptor Aly/REF (Yra1 in yeast) and the THO subcomplex (Katahira, 2012). The TREX/THO complex recruits the mRNA transport factor Nxf1-Nxt1 (TAP-p15; Mex67-Mtr2 in yeast) onto premRNA. The TREX2 complex is thought to facilitate mRNA export by the association of actively-transcribed genes with NPCs, a process known as “gene gating” (García-Oliver et al., 2012; Jani et al., 2014). The EJC, deposited on mRNA as a consequence of splicing, is supposed to constitute a binding platform for transiently associated factors, including the mRNA export factors Nxf1-Nxt1 and Aly/REF (Le Hir and Andersen, 2008). In addition, three shuttling SR proteins, SRSF1, SRSF3, and SRSF7, act as mRNA adaptors through their interaction with the cellular export factor Nxf1/TAP (Huang et al., 2003; Lai and Tarn, 2004; Hargous et al., 2006; reviewed in Zhong et al., 2009). XPO1/CRM1, through binding via adapter proteins, can mediate nuclear export of a subset of endogenous mRNA transcripts (for review, see Natalizio and Wente, 2013). Many mRNA export factors have been identified, and comparative genomics indicated that the proteins implicated in mRNA export are the less conserved factors across eukaryotes among the different RNA export pathways (Serpeloni et al., 2011). In plants, the regulation of mRNA export is far from being understood (Meier, 2012; Gaouar and Germain, 2013). Plant homologs of the mRNA transport adapter Nxf1-Nxt1 are not clearly identified and genes encoding those factors seem to be missing in plant genome (Serpeloni et al., 2011). Thus far, to the best of our knowledge, none of the plant SR proteins has been found to have a direct role in mRNA export activity. SR33 colocalizes in nuclear speckles with HPR1, a component of the THO complex required for RTE1 (Reversion-to-ethylene sensitivity 1 overexpressor) expression, but appears to have a role in transcription elongation rather than mRNA export (for review, see Xu et al., 2015a). Our results suggest the role of XPO1 in the active nuclear export of mRNAs and transiently associated SR proteins since the shuttling of distinct Arabidopsis SR proteins is significantly inhibited by LMB in transient as well as in stable expression assays. XPO1 mediates the nuclear export of proteins that possess Leu-rich NES sequences. Searching for NES motif in Arabidopsis SR proteins did not identify any export signal (Xu et al., 2015b). It is therefore very unlikely that they interact directly with XPO1. In plants, mRNA export might involve different pathways and the use of one of these could depend on the cellular state or mRNA nature (Rausin et al., 2010). Understanding the molecular mechanisms of how plant SR protein function in this essential process is challenging. Approaching such a complex problem requires a more global comprehensive view of SR protein and mRNA export complex interactome.
We demonstrate an interaction between SRSF1-subfamily SR proteins and SR45.1 and SR45.2, two isoforms resulting from SR45 alternative splicing. Our previous phylogenetic analyses showed that SR45 belongs to the plant SR family despite an atypical structural organization with a single RRM located between two distinct N- and C-terminal RS domains. We previously confirmed that SR45 is orthologous to animal and fungal RNPS1, a peripheral component of the EJC (Califice et al., 2012). During Arabidopsis development, SR45.1 plays a role in flower petal development and SR45.2 is required for normal root growth (Zhang and Mount, 2009). SR45 was recently found to negatively regulate Glc signaling during early Arabidopsis seedling development by down-regulating ABA signaling (Carvalho et al., 2010). The in vivo interactions of SRSF1-like proteins and SR45 seems to be supported by their concordant expression profiles during Arabidopsis development (this study and Zhang and Mount, 2009). The expression profiles of SR30, SR34, and SR34a were indeed very similar, though not strictly identical, and were observed mainly in growing tissues and metabolically active and dividing cells (i.e. root meristems, leaf primordia, pollen). Interaction between SR45 and SR34/SR34a had been recently detected by coimmunoprecipitation (Zhang et al., 2014). Earlier studies already identified several SR45-interacting splicing factors (SCL33, U1-70k, U2AF35, or SKIP; see Reddy, 2004; Day et al., 2012; Wang et al., 2012). Taken together, our observations furthermore argue that Arabidopsis SR proteins are closely interconnected and tethered to mRNA, not only through their function in splicing, but also as a postsplicing complex to promote/initiate mRNP nuclear export using a XPO1-dependent pathway. SR45(RNPS1), as a putative component of the EJC, might be at the heart of a complex interaction network having multiple role in mRNA processing into export-competent mRNP. Further functional studies will be required to elucidate the putative role of SR proteins (including SR45) in XPO1-dependent export of mRNP and to understand the processes of export complex assembly and mRNP structural remodeling during translocation through NPC. Intriguingly, our mutagenesis analysis showed that the mode of recognition of SR45 by SR34 involves the residues of the RNP motifs that contact RNA. The dual role of SR34 RRM1 also implies that protein interaction and RNA binding would be mutually exclusive. The RRM domain of some animal EJC components (Y14-Magoh) and EJC-associated NMD proteins (Upf3-Upf2) has been already shown to mediate protein-protein interactions through the β-sheet surface generally involved in RNA interactions (Shi and Xu, 2003; Kadlec et al., 2004). In addition, in Arabidopsis, SR45 was recently found to be required for the RNA-directed DNA methylation (Ausin et al., 2012). It has been recently proposed that the splicing machinery is involved in promoting RNA-directed DNA methylation and transcriptional silencing (Zhang et al., 2013). Whether plant SRSF1-like proteins are involved in these molecular and developmental processes is currently unknown.
MATERIALS AND METHODS
Plant Growth and Plant Transformation
Nicotiana tabacum (cv Petit Havana) and Arabidopsis (Arabidopsis thaliana) transient transformations by Agrobacterium infiltration were performed as described (Rausin et al., 2010).
Arabidopsis plants were stably transformed by floral dipping, and T3 homozygous lines were analyzed. For expression profiling, Arabidopsis plants (ecotype Col-0) were hydroponically grown from seeds in Hoagland medium as described (Talke et al., 2006). After six weeks of growth in a climate-controlled chamber at 21°C with a photoperiod of 16 h at a light intensity of 100 µmol m−2 s−1, root, rosette leave, cauline leave, inflorescence, and silique tissues were harvested separately from 12 plants. The tissues from the individual plants were pooled, frozen in liquid nitrogen, and stored at −80°C until further processing.
Binary Vector Constructions
All binary vector constructions were made using the pBI121 vector. All PCRs were carried out using Pfu polymerase (Promega) on Arabidopsis cDNA libraries or genomic DNA (Col-0 ecotype). A list of primers used in this study is provided in Supplemental Table S1. All constructs were verified by sequencing. All final plasmids were electroporated into the Agrobacterium tumefaciens strain GV3101 (pMP90) and subsequently used for plant transformations.
The construction of P35S:SR34-GFP in pBI121 was described in a previous report (Tillemans et al., 2005). The SR34a, SR34b, SR30, and SR45 cDNAs were cloned at the BamHI/KpnI sites of P35S:SR34-GFP to replace the SR34 cDNA.
From there, an identical cloning strategy has been used for the four SRSF1 genes (generically named here SR3x). The promoter regions of SR3x (∼ 1500 bp upstream of the ATG) were amplified by PCR from genomic DNA. The promoter amplicons were ligated at the HindIII/BamHI sites of P35S:SR3x-GFP vectors to replace the 35S promoter and create PSR3x:SR3x-GFP vectors, respectively. To obtain the PSR3x:GUS vectors, the pSR3x amplicons were cloned at the HindIII/BamHI sites of pBI121 upstream the GUS coding sequence, respectively. Note that for PSR34b, a NarI restriction site has been used for cloning instead of HindIII, as a restriction site for this enzyme is present in the PSR34b sequence.
The mutated SR34 versions were obtained by PCR-based site-directed mutagenesis on the SR34 coding sequence as described (Rausin et al., 2010). The three RS mutated domains were synthesized by GenScript and used as megaprimers to replace the wild-type RS domain by PCR in the SR34 cDNA (Geiser et al., 2001; Miyazaki et al., 2002), then processed as described (Rausin et al., 2010).
To generate BiFC constructs, the YFPC and YFPN fragments were amplified by PCR from the pBI121-35S:YFP vector (Rausin et al., 2010) and cloned into P35S:SR34-GFP and p35S:SR45-GFP at the KpnI/SacI sites to replace the GFP, respectively. The following linkers, RSIAT and RPACKIPNDLKQKVMNH, have been inserted between the SRxx cDNA and the YFPN or YFPC fragments, respectively, according to (Lu et al., 2010).
To generate FLIM-FRET constructs, the mCherry coding sequence was amplified from the pSAV047 vector (Fraipont et al., 2011) and cloned at the KpnI/SacI sites of the P35S:SR45-GFP vector to replace the GFP coding sequence. To generate the P35S:mCherry-GFP control, the amplicon was also ligated in P35S:SR34-GFP at the BamHI/KpnI sites to replace the SR34 coding sequence.
Yeast Two-Hybrid System
To generate the bait construct, SR34 wild-type and mutant cDNAs were cloned into the pGBKT7 vector (Clontech) at the EcoRI and BamHI restriction sites (see Supplemental Table S1 for primer list). The SR45 cDNA was cloned into the prey vector pGADT7-AD (Clontech) at the BamHI and SacI restriction sites.
The vectors and strains provided in the Matchmaker Gold Yeast Two-Hybrid System (Clontech) were used to screen a cDNA library from Arabidopsis (Mate and Plate Library-Universal Arabidopsis, Clontech) using SR34 as bait. Manipulation of the yeast cells and library screening were carried out according to the manufacturer’s instructions (Clontech). Briefly, the yeast (Saccharomyces cerevisiae) reporter strain Y2HGold was first transformed with the bait vector then crossed with strain Y187 containing the Arabidopsis cDNA library. After crossing, cells were plated onto a selective medium (−Trp/−Leu/X-α-gal/Aureobasidin A) to select resistant clones. Potential interactions were confirmed on a more selective medium (-Trp/-Leu/-Ade/-His/X-α-gal/Aureobasidin A) to increase the stringency of the screening. The prey plasmids were extracted from yeast cells (Easy Yeast Plasmid Isolation Kit (Clontech), cloned into E. coli, then prepared and sequenced.
For targeted interaction analysis, the cDNAs were cloned in frame in the multiple cloning site of pGBKT7 (SR30, SR34a) or pGADT7-AD (SR30, SR34a, CypRS64 and SRPK4; see Supplemental Table S1). Interactions were tested on selective medium (−Trp/−Leu/−Ade/−His/X-α-gal/Aureobasidin A).
RNA Extraction, cDNA Synthesis, and Quantitative RT-PCR
Total DNase-treated RNAs were extracted using the RNeasy plant mini kit and RNase-free DNase set (Qiagen). cDNAs were synthesized from 1.5 µg of total RNAs using oligo dT and the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas).
Quantitative RT-PCR reactions were performed as described earlier (Rausin et al., 2010) in 384-well plates with an ABI Prism 7900HT system (Applied Biosystems) using Maxima SYBR Green qPCR Master Mix (Fermentas) on material from three independent biological experiments, and a total of three technical repeats were run for each combination of cDNA and primer pair. The quality of the PCR reactions was checked visually through analysis of dissociation and amplification curves, and reaction efficiencies were determined for each PCR reaction using the LinRegPCR software (Ramakers et al., 2003). Mean reaction efficiencies were then determined for each primer pair from all reactions (>45 reactions; Supplemental Table S1) and used to calculate relative gene expression level by normalization using the reference gene At1g58050 with the qBase software (Hellemans et al., 2007). Four reference genes (UBQ10, EF1α, At1g58050, and At1g62930) were initially selected from the literature (Czechowski et al., 2005) and tested. Their adequacy to normalize gene expression in our experimental conditions was verified using the geNorm software (Vandesompele et al., 2002), and At1g58050 was identified as the best gene for normalization.
Analysis of GUS Reporter Lines
Histochemical GUS staining was carried out as described (Rausin et al., 2010) on Arabidopsis seedlings grown on 1/2 MS medium and on tissues of mature plants grown hydroponically. Harvested tissues were incubated in staining solution for 1 night to 2 d, then ethanol extracted and fixed before observation. Samples were observed under a Nikon SMZ1500 stereomicroscope equipped with a Nikon Digital Sight DS-U1 camera.
Confocal Microscopy, Photobleaching Experiments, FLIM, and Data Analysis
Leica TCS SP2 and SP5 inverted confocal laser microscopes (Leica Microsystems) were used for live cell imaging. The FLIP-shuttling experiments were carried out as previously described (Tillemans et al., 2006; Rausin et al., 2010).
Fluorescence lifetime measurements were performed on a SP5 SMD equipped with a time-correlated single photon counting module (PicoQuant GmbH) and a Chameleon pulsed infrared laser (tuned at 890 nm; Coherent). Photons were collected with either internal or external photomultiplier tubes. A water-immersion PlanApochromat 63×/NA objective (Leica) was used. Acquisition was performed using the LAS AF Version 2.4.1 (Leica) and SymphoTime Version 5.3 (PicoQuant GmbH) software. Briefly, the samples were continuously scanned for achieving sufficient photon statistics for the fitting of fluorescence decays, and data were analyzed using SymphoTime software. Fluorescence lifetimes of GFP (donor) were collected in nuclei showing colocalization of GFP and mCherry, and for each experiment, at least three nuclei were randomly selected for acquisition. In this work, we always performed the fitting on a single region of interest corresponding to the nucleoplasm of the cells in order to circumvent evident chlorophyll lifetime contribution in the analysis.
Fragments of transiently transformed leaves and stably transformed Arabidopsis roots were used for LMB treatments as previously described (Rausin et al., 2010). Briefly, LMB (stock solution at 5 µg/ml in 70% methanol; Sigma-Aldrich) was diluted in water and used at a final concentration of 10 nM. Plant cells were treated with LMB (or water as a control) for up to 2 h and were processed for imaging as described above. All observations and treatments were performed in at least three independent transient transformation events. The total number of analyzed nuclei for each experiment is mentioned in the main text. For statistical analysis, the fluorescence intensities at a given time point (50% of the time scale) of each FLIP-shuttling experiment were processed using GraphPad Prism, version 5 (GraphPad Software, Inc.). For statistical analysis of normality, the D’Agostino and Pearson test was used. To calculate the significance of the differences between fluorescence intensities, an unpaired t test (parametric data) was performed. When at least one of the series of data failed the normality test, the comparison between the experiments were performed with the Wilcoxon signed-rank test. The P values < 0.05 were considered to be a statistically significant difference.
Modeling
The NMR structure of the RRM of the human SRp20 bound to the RNA CAUC (pdb code: 2I2Y, Hargous et al., 2006) was used to model the structure of the SR34 RRM1 domain. Substitution of SRp20 side chains by those of SR34 was followed by an energy minimization of the structure with the program Yasara (Krieger et al., 2004), using a standard protocol consisting in a steepest descent minimization followed by simulated annealing. Minimization parameters consisted in the use of Yasara2 force field (Krieger et al., 2009), a cutoff distance of 7.86 Å, particle mesh Ewald, long range electrostatics (Essmann et al., 1995), periodic boundary conditions, and water-filled simulation cell. The structure of SR34 RRM2 domain was modeled, using the same protocol, from the solution structure of the human SRSF1 pseudo-RRM bound to RNA (pdb code: 2M8D, Cléry et al., 2013). The nucleotides were not used in minimizations.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_172386 (SR30), NP_850933(SRp34), NP_190512 (SR34a), NP_567235 (SRp34b).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression profiles SRSF1 members and localization of GFP translational fusions.
Supplemental Figure S2. Diagram depicting the structure of wild-type SR34 and generated mutant derivatives.
Supplemental Figure S3. Modeling of the SR34 RRM1 domain
Supplemental Figure S4. Modeling of the SR34 RRM2 domain
Supplemental Figure S5. FLIP-shuttling of SR34rnp1 in Arabidopsis cells.
Supplemental Figure S6. Identification of SR34 mutations that influence interaction with SR45 and SRPK4 in an Y2H assay.
Supplemental Figure S7. In vivo detection of protein-protein interaction between SR34 and SR45 by BiFC and FLIM-FRET.
Supplemental Figure S8. FLIP-shuttling of SR45.
Supplemental Table S1. List of primers used in this study
Supplementary Material
Acknowledgments
We thank Gentiane Haesbroeck (Department of Mathematics, ULg) for helpful advices on statistical analysis, Steven Fanara for discussions, and Dr. Nicolas Dony for plamid pSAV047.
Glossary
- NPC
nuclear pore complex
- NLS
nuclear localization signals
- NES
nuclear export signal
- TRN-SR
Transportin-SR
- XPO1
Exportin-1
- EJC
exon junction complex
- FLIP
Fluorescence Loss In Photobleaching
- LMB
leptomycin B
- BiFC
bimolecular fluorescence complementation
- FRET
fluorescence resonance energy transfer
- FLIM
fluorescence lifetime imaging
References
- Änkö ML. (2014) Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol 32: 11–21 [DOI] [PubMed] [Google Scholar]
- Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA (2013) Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J Mol Biol 425: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin I, Greenberg MV, Li CF, Jacobsen SE (2012) The splicing factor SR45 affects the RNA-directed DNA methylation pathway in Arabidopsis. Epigenetics 7: 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Lorković ZJ (2008) Plant SR proteins and their functions. Curr Top Microbiol Immunol 326: 83–102 [DOI] [PubMed] [Google Scholar]
- Barta A, Kalyna M, Reddy AS (2010) Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22: 2926–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Zhou X, Meier I (2012) Dynamics of the plant nuclear envelope and nuclear pore. Plant Physiol 158: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR (1993) Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR (1997) Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol 138: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califice S, Baurain D, Hanikenne M, Motte P (2012) A single ancient origin for prototypical serine/arginine-rich splicing factors. Plant Physiol 158: 546–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardarelli F, Lanzano L, Gratton E (2012) Capturing directed molecular motion in the nuclear pore complex of live cells. Proc Natl Acad Sci USA 109: 9863–9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RF, Carvalho SD, Duque P (2010) The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiol 154: 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF (2002) Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol 22: 6871–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléry A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–298 [DOI] [PubMed] [Google Scholar]
- Cléry A, Sinha R, Anczuków O, Corrionero A, Moursy A, Daubner GM, Valcárcel J, Krainer AR, Allain FH (2013) Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc Natl Acad Sci USA 110: E2802–E2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Cléry A, Allain FH (2013) RRM-RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol 23: 100–108 [DOI] [PubMed] [Google Scholar]
- Day IS, Golovkin M, Palusa SG, Link A, Ali GS, Thomas J, Richardson DN, Reddy AS (2012) Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing. Plant J 71: 936–947 [DOI] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Anrather D, Roitinger E, Djamei A, Hufnagl T, Barta A, Csaszar E, Dohnal I, Lecourieux D, Hirt H (2006) Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res 34: 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8593 [Google Scholar]
- Fang Y, Hearn S, Spector DL (2004) Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol Biol Cell 15: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilner T, Hultschig C, Lee J, Meyer S, Immink RG, Koenig A, Possling A, Seitz H, Beveridge A, Scheel D, Cahill DJ, Lehrach H, et al. (2005) High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol Cell Proteomics 4: 1558–1568 [DOI] [PubMed] [Google Scholar]
- Field MC, Koreny L, Rout MP (2014) Enriching the pore: splendid complexity from humble origins. Traffic 15: 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraipont C, Alexeeva S, Wolf B, van der Ploeg R, Schloesser M, den Blaauwen T, Nguyen-Distèche M (2011) The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157: 251–259 [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Carvalho MP, Kehlenbach A, Valcarcel J, Carmo-Fonseca M (2001) Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J Biol Chem 276: 13104–13112 [DOI] [PubMed] [Google Scholar]
- Gaouar O, Germain H (2013) mRNA export: threading the needle. Front Plant Sci 4: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Oliver E, García-Molinero V, Rodríguez-Navarro S (2012) mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta 1819: 555–565 [DOI] [PubMed] [Google Scholar]
- Geiser M, Cebe R, Drewello D, Schmitz R (2001) Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31: 88–90, 92 [DOI] [PubMed] [Google Scholar]
- Goryaynov A, Ma J, Yang W (2012) Single-molecule studies of nucleocytoplasmic transport: from one dimension to three dimensions. Integr Biol (Camb) 4: 10–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH (2006) Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Baker BS (1997) In vivo analysis of the functional domains of the Drosophila splicing regulator RBP1. Proc Natl Acad Sci USA 94: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stévenin J, Steitz JA (2003) SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell 11: 837–843 [DOI] [PubMed] [Google Scholar]
- Jani D, Valkov E, Stewart M (2014) Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res 42: 6686–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S (2004) The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 11: 330–337 [DOI] [PubMed] [Google Scholar]
- Kalyna M, Barta A (2004) A plethora of plant serine/arginine-rich proteins: redundancy or evolution of novel gene functions? Biochem Soc Trans 32: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. (2012) mRNA export and the TREX complex. Biochim Biophys Acta 1819: 507–513 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14: 153–165 [DOI] [PubMed] [Google Scholar]
- Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G (2004) Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57: 678–683 [DOI] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77(Suppl 9): 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci USA 98: 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Tarn WY (2004) Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem 279: 31745–31749 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR (2008) Structural insights into the exon junction complex. Curr Opin Struct Biol 18: 112–119 [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorković ZJ, Barta A (2002) Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J Biol Chem 277: 39989–39998 [DOI] [PubMed] [Google Scholar]
- Lopato S, Kalyna M, Dorner S, Kobayashi R, Krainer AR, Barta A (1999) atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev 13: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Hilscher J, Barta A (2008) Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp Cell Res 314: 3175–3186 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Lopato S, Pexa M, Lehner R, Barta A (2004) Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleoprotein-specific proteins suggest their involvement in pre-mRNA Splicing. J Biol Chem 279: 33890–33898 [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EW, Harada JJ, Rothstein SJ, Cui Y (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61: 259–270 [DOI] [PubMed] [Google Scholar]
- Maertens GN, Cook NJ, Wang W, Hare S, Gupta SS, Öztop I, Lee K, Pye VE, Cosnefroy O, Snijders AP, KewalRamani VN, Fassati A, et al. (2014) Structural basis for nuclear import of splicing factors by human Transportin 3. Proc Natl Acad Sci USA 111: 2728–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Krainer AR (2010) A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24: 1073–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I. (2012) mRNA export and sumoylation-Lessons from plants. Biochim Biophys Acta 1819: 531–537 [DOI] [PubMed] [Google Scholar]
- Merkle T. (2003) Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signalling. Curr Genet 44: 231–260 [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Takenouchi M (2002) Creating random mutagenesis libraries using megaprimer PCR of whole plasmid. Biotechniques 33: 1033–1034, 1036–1038 [DOI] [PubMed] [Google Scholar]
- Natalizio BJ, Wente SR (2013) Postage for the messenger: designating routes for nuclear mRNA export. Trends Cell Biol 23: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rausin G, Tillemans V, Stankovic N, Hanikenne M, Motte P (2010) Dynamic nucleocytoplasmic shuttling of an Arabidopsis SR splicing factor: role of the RNA-binding domains. Plant Physiol 153: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AS. (2004) Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci 9: 541–547 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Day IS, Göhring J, Barta A (2012) Localization and dynamics of nuclear speckles in plants. Plant Physiol 158: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpeloni M, Vidal NM, Goldenberg S, Avila AR, Hoffmann FG (2011) Comparative genomics of proteins involved in RNA nucleocytoplasmic export. BMC Evol Biol 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR (2004) Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell 13: 367–376 [DOI] [PubMed] [Google Scholar]
- Shi H, Xu RM (2003) Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev 17: 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151: 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talke IN, Hanikenne M, Krämer U (2006) Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol 142: 148–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Hara-Nishimura I (2014) Functional insights of nucleocytoplasmic transport in plants. Front Plant Sci 5: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillemans V, Dispa L, Remacle C, Collinge M, Motte P (2005) Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J 41: 567–582 [DOI] [PubMed] [Google Scholar]
- Tillemans V, Leponce I, Rausin G, Dispa L, Motte P (2006) Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors. Plant Cell 18: 3218–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, Wilson SA (2007) Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep 8: 756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman. F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, Jiao Y, Puta F, et al. (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhou X, Wen CK (2015a) HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet 11: e1004956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Marquis K, Pei J, Fu SC, Cağatay T, Grishin NV, Chook YM (2015b) LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 31: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Zhang Z, Jing B, Gannon P, Ding J, Xu F, Li X, Zhang Y (2011) Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet 7: e1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CY, Velazquez-Dones AL, Lyman SK, Fu XD (2003) Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J Biol Chem 278: 18050–18055 [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Zhou JX, Liu J, Ma ZY, Zhang SW, Dou K, Huang HW, Cai T, Liu R, Zhu JK, He XJ (2013) The splicing machinery promotes RNA-directed DNA methylation and transcriptional silencing in Arabidopsis. EMBO J 32: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XN, Mo C, Garrett WM, Cooper B (2014) Phosphothreonine 218 is required for the function of SR45.1 in regulating flower petal development in Arabidopsis. Plant Signal Behav: e29134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XN, Mount SM (2009) Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol 150: 1450–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD (2009) SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.