Nitrate activates a set of glutaredoxin proteins that act to slow the growth of the root.
Abstract
Nitrogen is an essential soil nutrient for plants, and lack of nitrogen commonly limits plant growth. Soil nitrogen is typically available to plants in two inorganic forms: nitrate and ammonium. To better understand how nitrate and ammonium differentially affect plant metabolism and development, we performed transcriptional profiling of the shoots of ammonium-supplied and nitrate-supplied Arabidopsis (Arabidopsis thaliana) plants. Seven genes encoding class III glutaredoxins were found to be strongly and specifically induced by nitrate. RNA silencing of four of these glutaredoxin genes (AtGRXS3/4/5/8) resulted in plants with increased primary root length (approximately 25% longer than the wild type) and decreased sensitivity to nitrate-mediated inhibition of primary root growth. Increased primary root growth is also a well-characterized phenotype of many cytokinin-deficient plant lines. We determined that nitrate induction of glutaredoxin gene expression was dependent upon cytokinin signaling and that cytokinins could activate glutaredoxin gene expression independent of plant nitrate status. In addition, crosses between “long-root” cytokinin-deficient plants and “long-root” glutaredoxin-silenced plants generated hybrids that displayed no further increase in primary root length (i.e. epistasis). Collectively, these findings suggest that AtGRXS3/4/5/8 operate downstream of cytokinins in a signal transduction pathway that negatively regulates plant primary root growth in response to nitrate. This pathway could allow Arabidopsis to actively discriminate between different nitrogen sources in the soil, with the preferred nitrogen source, nitrate, acting to suppress primary root growth (vertical dimension) in concert with its well-characterized stimulatory effect on lateral root growth (horizontal dimension).
Most plants are wholly dependent on soil reserves of inorganic or organic nitrogen to support their growth. As a structural component of chlorophyll, proteins, and nucleic acids, nitrogen is required in larger amounts than any other mineral nutrient. Thus, most current agricultural practices call for substantial inputs of chemical fertilizers that are rich in inorganic nitrogen, typically in the form of nitrate (NO3−) and/or ammonium (NH4+). Nitrate and ammonium have different soil retention properties, with ammonium typically being adsorbed to negatively charged soil particles, while nitrate leaches from agricultural soils relatively quickly, potentially contributing to eutrophication of aquatic systems (Gruber and Galloway, 2008). Likewise, nitrate and ammonium can have differential effects on plant physiology and development. Most plants display a clear preference for one form of inorganic nitrogen, though substantial differences can exist among cultivars/ecotypes of a single species (Boudsocq et al., 2012; Sarasketa et al., 2014). Although ammonium requires less reductant to assimilate, nitrate is the preferred nitrogen source for many crop species, and the provision of ammonium as the sole source of nitrogen can be toxic (Britto and Kronzucker, 2002; Li et al., 2014). From a physiological perspective, nitrate nutrition is associated with elevated cytokinin biosynthesis, increased organic acid content, and soil alkalinization, while ammonium nutrition is associated with increased activity of alternative respiratory pathways, elevated amino acid content, and soil acidification (Walch-Liu et al., 2005; Patterson et al., 2010). In addition, recent studies have demonstrated distinct transcriptome, proteome, and protein phosphorylation signatures in plants supplied with either ammonium or nitrate as a nitrogen source (Patterson et al., 2010; Møller et al., 2011; Engelsberger and Schulze, 2012).
In this study, we have compared global transcriptional responses to nitrate and ammonium in Arabidopsis (Arabidopsis thaliana), leading to the identification of a group of class III glutaredoxin genes that are specifically responsive to plant nitrate status. Several of the nitrate-induced glutaredoxins act as negative regulators of primary root growth. Further, they appear to act downstream of cytokinins in a common signal transduction pathway. This pathway, which regulates primary root growth, may work in concert with well-characterized lateral root growth developmental programs, modulating root system architecture to maximize plant utilization of nitrate-rich regions of the soil.
RESULTS
Comparative Transcriptomics of the Shoots of Nitrate- and Ammonium-Supplied Plants
To complement our previous studies on the effects of nitrate and ammonium on the Arabidopsis root transcriptome (Patterson et al., 2010), hydroponically grown Arabidopsis plants were subjected to a brief (26 h) nitrogen starvation treatment and then transferred to media containing either 1 mm nitrate or 1 mm ammonium as the sole nitrogen source. Shoot tissue was harvested after 8 h and RNA was isolated for microarray analysis (Affymetrix ATH1 GeneChip). Compared to “time 0” (just prior to nitrogen addition), 1,127 genes showed differential expression in ammonium-treated plants, and 1,383 genes showed differential expression in nitrate-treated plants. There was substantial overlap (>66%) between the nitrate-regulated and ammonium-regulated gene sets, with these “shared” genes likely representing general responses to the resupply of nitrogen in a nitrogen-limited plant. In contrast, a relatively small number of genes showed a robust “nitrate-specific” response (103 genes) or a robust “ammonium-specific” response (50 genes) in shoots (Supplemental Tables S1 and S2). Nitrate-specific and ammonium-specific transcriptional responses in shoots showed little overlap with the previously described nitrate- and ammonium-specific transcriptional responses in roots (Patterson et al., 2010), suggesting substantial organ specificity in unique physiological responses to nitrate and ammonium (Supplemental Fig. S1).
Analysis of nitrate- and ammonium-specific transcriptional responses using the BioMaps program (Katari et al., 2010) revealed several overrepresented functional categories (Table I). Most notably, genes associated with “cytokinin response,” “oxygen and radical detoxification,” and “detoxification by modification” categories were strongly overrepresented in the nitrate-specific shoot response. The nitrate-cytokinin association is unsurprising given the well-established relationship among nitrate nutrition, cytokinin biosynthesis, and shoot growth (Walch-Liu et al., 2005; Sakakibara et al., 2006). However, the connection between nitrate and a putative oxidative stress response is less obvious. Nitrate and ammonium did not substantially affect hydrogen peroxide levels or oxidative damage in shoots (Supplemental Fig. S2), suggesting that neither of the treatments place the shoot under oxidative stress.
Table I. Overrepresented functional categories in nitrate-specific and ammonium-specific transcriptional responses.
| Functional Category | Observed Frequency | Expected Frequency | P Value |
|---|---|---|---|
| Induced by nitrate | |||
| Detoxification by modification | 10.7% | 0.1% | 4.05E-11 |
| Cytokinin response | 8.9% | 0.1% | 1.27E-06 |
| Cell growth/morphogenesis | 12.5% | 0.9% | 3.60E-05 |
| Oxygen and radical detoxification | 12.5% | 0.9% | 5.83E-05 |
| Electron transport | 16.1% | 2.5% | 5.50E-04 |
| Repressed by nitrate | |||
| Metabolism of stilbenes, flavonoids | 6.5% | 0.2% | 2.48E-03 |
| Oxygen and radical detoxification | 10.9% | 0.9% | 3.68E-03 |
| Induced by ammonium | |||
| Lipid binding | 16.7% | 0.5% | 1.13E-05 |
| Lipid/fatty acid transport | 10.0% | 0.4% | 8.73E-03 |
| Repressed by ammonium | |||
| Cellular sensing and response to external stimulus | 42.1% | 4.7% | 5.18E-05 |
| Nitrogen, sulfur, and selenium metabolism | 15.8% | 0.6% | 7.87E-03 |
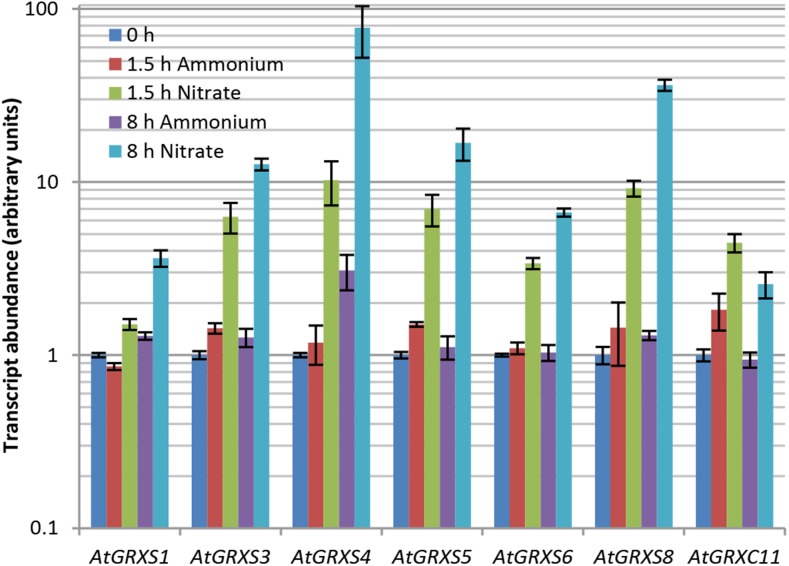
A closer examination of the “detoxification by modification” category and its subcategory “oxygen and radical detoxification” revealed that most of the differentially expressed genes identified in these categories encode glutaredoxin proteins. Further, within the nitrate-specific gene set, the “top five” genes showing the largest quantitative increase in transcript abundance were all glutaredoxins (Supplemental Table S1). Subsequent real-time reverse transcription PCR (RT-PCR) analyses of RNA isolated from the shoots of hydroponically grown Arabidopsis plants confirmed a rapid, sustained, and nitrate-specific up-regulation of the glutaredoxin genes AtGRXS1 (At1g03020), AtGRXS3 (At4g15700), AtGRXS4 (At4g15680), AtGRXS5 (At4g15690), AtGRXS6 (At3g62930), AtGRXS8 (At4g15660), and AtGRXC11 (At3g62950; Fig. 1). Of these seven glutaredoxins, only AtGRXS6 was identified as nitrate regulated in our previous microarray analysis of nitrate- and ammonium-treated root tissue (Patterson et al., 2010). In addition, all of these genes show substantially higher basal expression in shoot tissue than in root tissue (Schmid et al., 2005; Winter et al., 2007; Belin et al., 2015). However, real-time RT-PCR analysis of RNA isolated from roots revealed a similar pattern of nitrate-specific induction for AtGRXS3, -4, -5, -6, and -8 (Supplemental Fig. S3). Thus, it appears that this group of glutaredoxins is globally up-regulated by nitrate, and could potentially play a role in tailoring the metabolism or development of Arabidopsis in response to this specific form of inorganic nitrogen.
Figure 1.
Nitrate-specific up-regulation of class III glutaredoxins in Arabidopsis shoots. Transcript abundance of the denoted glutaredoxin genes was measured by real-time RT-PCR in nitrate-treated and ammonium-treated plants. Data points represent means ± se (n = 3). Note that transcript abundance at time 0 h was arbitrarily assigned a value of 1.0, independently for each gene.
Functional Analyses of Nitrate-Regulated Glutaredoxins
Glutaredoxins are small oxidoreductase proteins that can reduce disulfide bonds and Cys-glutathione bonds (glutathione adducts) within target proteins (Meyer et al., 2009). Because Cys residues are commonly oxidized by reactive oxygen species, glutaredoxins are often considered to act as antioxidants that help to minimize oxidative damage to proteins. Based on the sequence characteristics of their active site motifs, glutaredoxins have been grouped into three classes (I–III). Class III glutaredoxins (also known as CC-type glutaredoxins) contain two consecutive cysteines in their active site motif and are exclusively found in higher plants (Ströher and Millar, 2012). The seven glutaredoxin genes identified as strongly and specifically induced by nitrate in our data are all class III. Only four of the 21 class III glutaredoxin genes in the Arabidopsis genome have been functionally characterized (Xing et al., 2005; Xing and Zachgo, 2008; La Camera et al., 2011; Laporte et al., 2012), and no functional information is available about any of the identified nitrate-induced glutaredoxins.
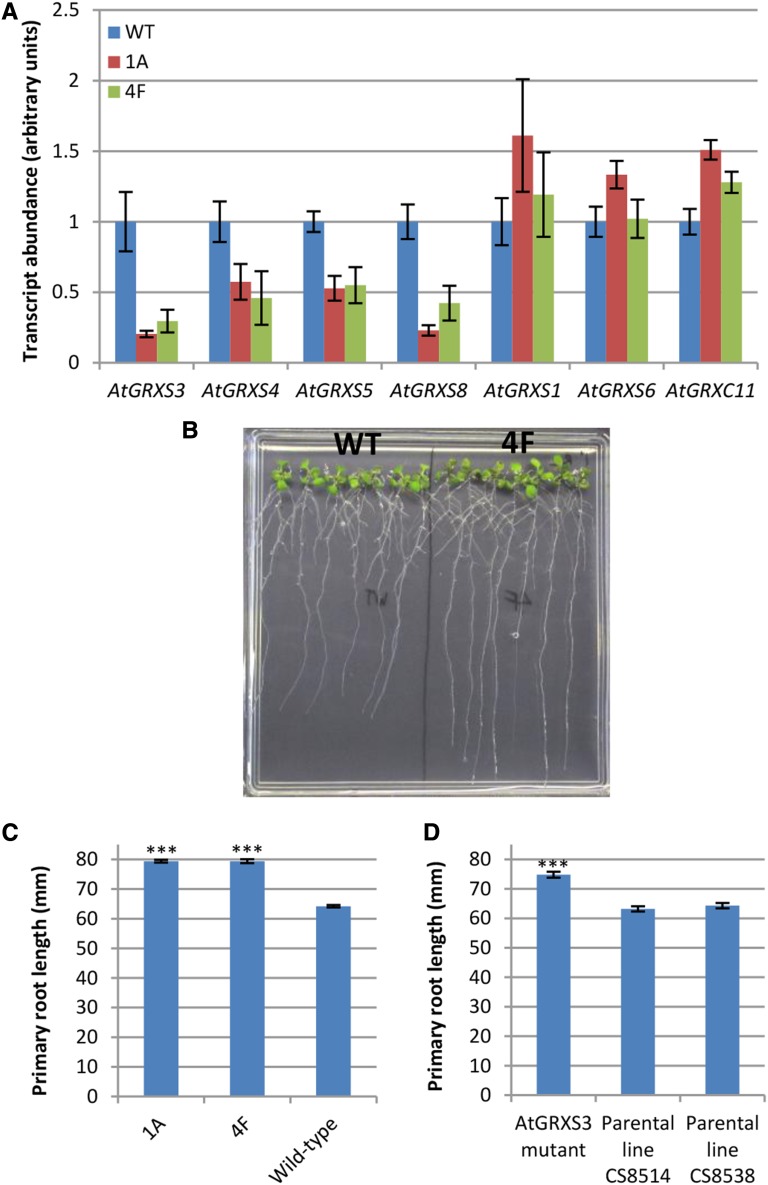
To gain some insight into the physiological function of these genes, we focused our initial studies on AtGRXS3, AtGRXS4, AtGRXS5, and AtGRXS8. These four genes are organized in a tandem array on Arabidopsis chromosome 4 and are extremely similar in sequence (Supplemental Fig. S4A). In addition, coexpression analyses using GeneCAT (Mutwil et al., 2008) demonstrated that these four genes form a tight coexpression network (Supplemental Fig. S4B). Thus, it is likely that AtGRXS3, AtGRXS4, AtGRXS5, and ATGRXS8 have arisen by relatively recent gene duplication events and that they display substantial regulatory and functional redundancy. We generated an RNA interference vector based on the sequence of AtGRXS3, which is >89% identical at the nucleotide sequence level to AtGRXS4, AtGRXS5, and AtGRXS8. Two of the transgenic Arabidopsis lines (1A and 4F) generated using this vector displayed substantial reductions in the transcript abundance of the target glutaredoxins (AtGRXS3, AtGRXS4, AtGRXS5, and AtGRXS8) without repressing expression of the other nitrate-regulated glutaredoxins (AtGRXS1, AtGRXS6, and AtGRXC11; Fig. 2A). Despite the generation of two affinity-purified peptide antibodies targeting different regions of the AtGRXS3 protein, none of the corresponding glutaredoxin proteins could be detected by immunoblotting. This finding is consistent with the absence of protein abundance data in previous studies of class III glutaredoxins (Xing et al., 2005; Xing and Zachgo, 2008; La Camera et al., 2011; Laporte et al., 2012) and in large-scale nontargeted proteomics studies (Baerenfaller et al., 2008), likely reflecting the very low abundance of these proteins in vivo (Ströher and Millar, 2012).
Figure 2.
Characterization of glutaredoxin-silenced plant lines. A, Transcript abundance of the noted glutaredoxin genes in wild-type (WT; Col-0) and AtGRXS3 RNA interference lines (1A and 4F). Compared to the other included glutaredoxin genes, AtGRXS1, AtGRXS6, and AtGRXC11 have weak sequence similarity to AtGRXS3, and are included as a negative controls. Data points represent means ± se (n = 6). B, Eleven-day-old wild-type and 4F seedlings grown on vertically oriented plates. C, Primary root length of 11-d-old seedlings grown on vertically oriented plates. Data points represent means ± se (n ≥ 36). D, Primary root length of 10-d-old seedlings of an AtGRXS3 mutant (ecotype Nössen) and its parental lines. Data points represent means ± se (n ≥ 39). A triple asterisk (***) indicates a significant difference compared to the control(s) with a P value ≤ 0.001.
When transgenic lines 1A and 4F were grown on soil, they displayed no consistent phenotypic differences from the wild type (Columbia-0) other than a slight delay in bolting. However, growth of 1A and 4F seedlings on vertically oriented plates revealed that they had significantly longer primary roots than wild-type plants (Fig. 2, B and C). In contrast to the clear primary root phenotype, no difference in the density or total length of lateral roots was observed in the glutaredoxin-silenced lines. An insertional mutant line was available for AtGRXS3 but not for the other glutaredoxins (presumably due to the small size of their coding sequences; 309 bp). The AtGRXS3 mutant line is in the Nössen ecotype of Arabidopsis, and it contains a Ds transposon in the middle of the gene coding sequence (Ito et al., 2005). A comparison of the AtGRXS3 mutant and its two parental lines again revealed a significantly longer primary root (Fig. 2D), though the relative root length of the AtGRXS3 mutant (1.17) was less than the relative root length of lines 1A and 4F (approximately 1.25), potentially reflecting some functional redundancy of AtGRXS4, AtGRXS5, and AtGRXS8 in the single mutant line. Overall, these findings suggest that these nitrate-induced glutaredoxins act as negative regulators of primary root growth.
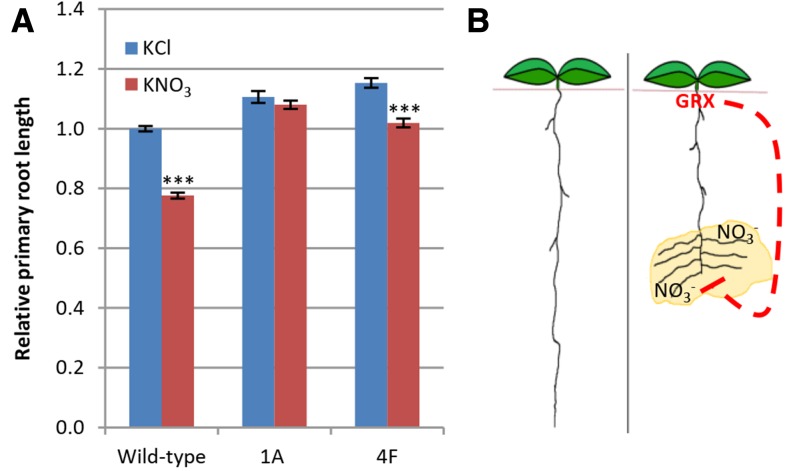
Previous research has shown that nitrate acts as a potent inhibitor of primary root growth in Arabidopsis plants grown hydroponically with ammonium as their primary nitrogen source (Vidal et al., 2010). As expected, when wild-type plants were grown under these conditions, the addition of nitrate to the medium substantially slowed primary root growth, with a 22.4% reduction in primary root length (Fig. 3A). However, glutaredoxin-silenced lines 1A and 4F were less sensitive to nitrate-mediated inhibition of primary root growth, with nitrate treatment decreasing root length of the 1A and 4F lines by 2.4% and 11.6%, respectively (Fig. 3A). The AtGRXS3 mutant line also demonstrated minimal nitrate-mediated inhibition of root growth in this system, but this was also true of its parental lines, suggesting that the Nössen ecotype is substantially less sensitive to nitrate-mediated inhibition of primary root growth than the Columbia-0 ecotype (Supplemental Fig. S5). Collectively, these findings suggest that the AtGRXS3/4/5/8 genes act to integrate information about plant nitrogen source with corresponding developmental programs controlling primary root development, as outlined in the model presented in Figure 3B.
Figure 3.
Glutaredoxins mediate the inhibitory effect of nitrate on primary root growth. A, Relative primary root length of 12-d-old wild-type and glutaredoxin-silenced (1A and 4F) plants. Plants were grown for 9 d in basal ammonium succinate medium and then 5 mm KNO3 or KCl was added to the medium. After 3 d, root length was measured. Data were combined from three separate experiments, with raw root length data normalized individually for each experiment by setting the average root length of wild-type plants supplied with KCl equal to 1.0. Data points represent means ± se (n ≥ 117). Asterisks indicate significant differences (P ≤ 0.001) compared to KCl-treated plants of the same genotype. Nitrate-associated reductions in primary root length are 22.4%, 2.4%, and 11.6% in wild-type, 1A, and 4F lines, respectively. B, Model outlining a role for glutaredoxins (GRX) in suppressing primary root growth in nitrate-rich regions of the soil. The dashed line represents glutaredoxin-initiated signaling that causes suppression of primary root growth. While primary root growth would be limited via this pathway, lateral root growth would be stimulated via an independent pathway (Zhang and Forde, 1998).
Connections Between Nitrate-Regulated Glutaredoxins and Cytokinin Signaling
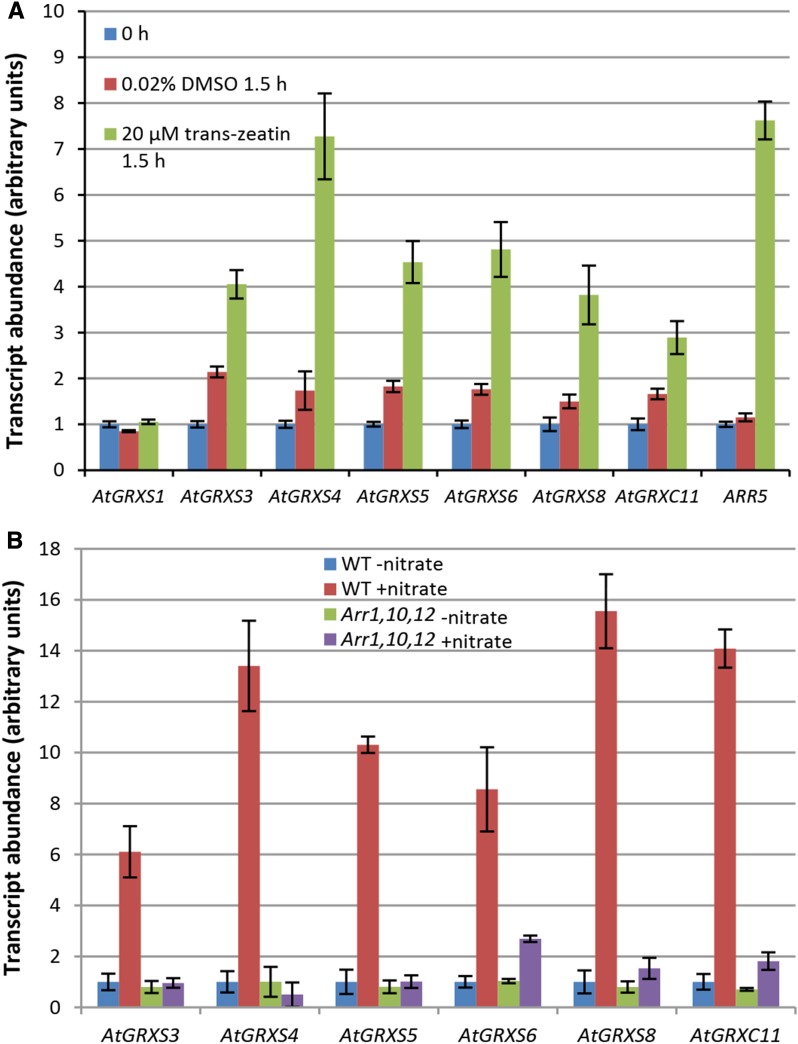
Because nitrate strongly up-regulates cytokinin biosynthesis in roots as well as the subsequent root-to-shoot transport of cytokinins, there can be extensive overlap between nitrate-regulated transcriptional responses and cytokinin-regulated transcriptional responses (Sakakibara et al., 2006). A comparison between transcriptome data derived from cytokinin-treated plants (Brenner et al., 2005; Kiba et al., 2005) and transcriptome data derived from our nitrate-treated plants showed relatively limited overlap (Supplemental Fig. S6), with the notable exceptions of AtGRXS4, AtGRXS5, AtGRXS6, and AtGRXC11, which were strongly induced by both treatments. To further explore the potential role of cytokinins in regulating glutaredoxin gene expression, hydroponically grown Arabidopsis plants were nitrogen starved for 26 h (as in previous experiments) and then the shoots were sprayed with a 20 µm trans-zeatin solution, without supplying nitrogen in any form. As shown in Figure 4A, all of the previously identified nitrate-induced glutaredoxin genes except AtGRXS1 were up-regulated by trans-zeatin, independent of plant nitrate status. Data mining of previously published transcriptome datasets using Genevestigator (Hruz et al., 2008) provided additional evidence of glutaredoxin gene regulation by cytokinins. Most notably, the cytokinin-deficient transgenic Arabidopsis line 35S:CKX1 showed strongly reduced expression of AtGRXS5, AtGRXS6, and AtGRXS8 (Brenner et al., 2005). In addition, AtGRXS6 and AtGRXC11 have been identified among 23 direct target genes of ARR1, one of the most extensively studied cytokinin response regulator transcription factors (Taniguchi et al., 2007).
Figure 4.
Regulation of class III glutaredoxins by cytokinin. A, Treatment of the shoots of hydroponically grown Arabidopsis plants with the cytokinin trans-zeatin. Transcript abundance of denoted glutaredoxin genes was measured by real-time RT-PCR. ARR5 is a cytokinin response regulator gene and serves as a positive control. Data points represent means ± se (n = 6). Trans-zeatin was dissolved in 0.02% dimethyl sulfoxide (DMSO). Transcript abundance at time 0 h was arbitrarily assigned a value of 1.0, independently for each gene. B, Regulation of class III glutaredoxins in the ARR1,10,12 cytokinin signaling mutant. Hydroponically grown wild-type and ARR1,10,12 plants were nitrogen starved for 26 h and then resupplied with 1 mm KNO3 (+nitrate) or 0.5 mm K2SO4 (−nitrate) for 8 h. Transcript abundance of the denoted glutaredoxin genes was measured by real-time RT-PCR. Data points represent means ± se (n = 3). Transcript abundance data for wild-type plants supplied with K2SO4 (WT −nitrate treatment) was arbitrarily assigned a value of 1.0, independently for each gene.
To determine whether cytokinin signaling is necessary for nitrate-mediated induction of glutaredoxin gene expression, we made use of the ARR1,10,12 mutant, which is deficient in cytokinin signal transduction. The ARR1, ARR10, and ARR12 proteins are a group of functionally redundant type-B response regulators (transcription factors) that control the expression of most cytokinin-regulated genes (Argyros et al., 2008; Ishida et al., 2008). When ARR1,10,12 mutants were grown hydroponically, nitrogen-starved, and then supplied with nitrate, the induction of AtGRXS3, AtGRXS4, AtGRXS5, AtGRXS6, AtGRXS8, and AtGRXC11 was almost completely abolished (Fig. 4B). Thus, it appears that cytokinin is both necessary and sufficient for nitrate-associated regulation of these glutaredoxin genes.
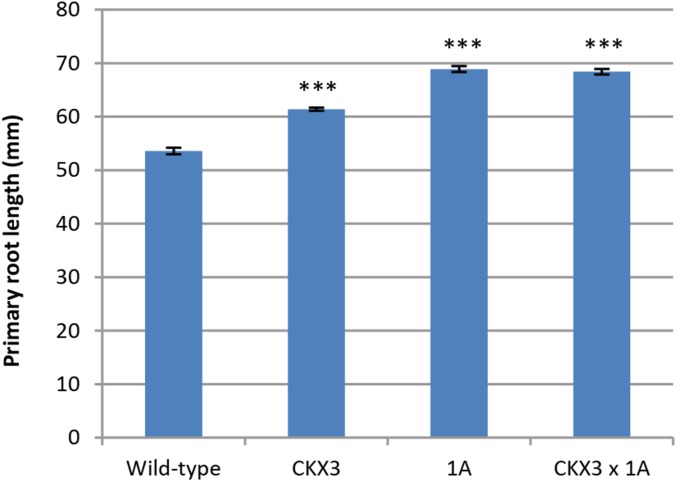
The regulation of AtGRXS3/4/5/8 by cytokinins is especially interesting in light of the shared primary root phenotype of glutaredoxin-silenced and cytokinin-deficient plant lines. Increased primary root length is a defining characteristic of many plants deficient in cytokinin synthesis or cytokinin signaling (e.g. ipt cytokinin biosynthesis mutants, 35S:CKX cytokinin-oxidase overexpressing lines, AHK cytokinin receptor mutants, and ARR cytokinin response regulator mutants; Hwang et al., 2012). Cytokinin-deficient plants also display several other typical phenotypic characteristics, including increased lateral root growth, reduced shoot growth, and increased seed size (Werner et al., 2003; Riefler et al., 2006), but none of these additional phenotypes was observed in glutaredoxin-silenced lines. Thus, AtGRXS3/4/5/8 are regulated by cytokinins, and silencing of these genes results in a phenocopy of the “long primary root” phenotype (but not the other phenotypes) observed in cytokinin-deficient plants. Examination of the root tips of glutaredoxin-silenced plants using confocal microscopy demonstrated that they also have an increased number of cortical cells in their primary root meristems (Supplemental Fig. S7), which is an additional characteristic of cytokinin-deficient lines (Dello Ioio et al., 2007). Like glutaredoxin-silenced plants, the cytokinin-deficient transgenic line 35S:CKX3 also displays decreased sensitivity to nitrate-mediated inhibition of primary root growth (Supplemental Fig. S5). Overall, these findings suggest that glutaredoxins might act as intermediates in a cytokinin-associated signal transduction pathway that specifically regulates primary root growth. To test this hypothesis, we utilized the 35S:CKX3 line, which overexpresses the catabolic gene cytokinin oxidase3, resulting in substantially decreased cytokinin levels and increased primary root growth on vertically oriented plates (Werner et al., 2003). As shown in Figure 5, crosses between the “long root” glutaredoxin-silenced line 1A and the “long root” 35S:CKX3 line produced progeny that displayed no additional increase in primary root growth compared to the parental lines. This epistatic effect further supports the hypothesis that glutaredoxins and cytokinins operate in a common signaling pathway that regulates primary root length in Arabidopsis.
Figure 5.
Epistatic effects of cytokinin and glutaredoxins in regulating primary root growth. Analyses were performed on 11-d-old seedlings grown on vertical plates. CKX3 is the 35S:CKX3 cytokinin-deficient transgenic line, 1A is a glutaredoxin-silenced transgenic line, and CKX3 × 1A is a cross between these two lines. All data points represent means ± se (n ≥ 22). A triple asterisk (***) indicates a significant difference compared to the wild type with a P value ≤ 0.001.
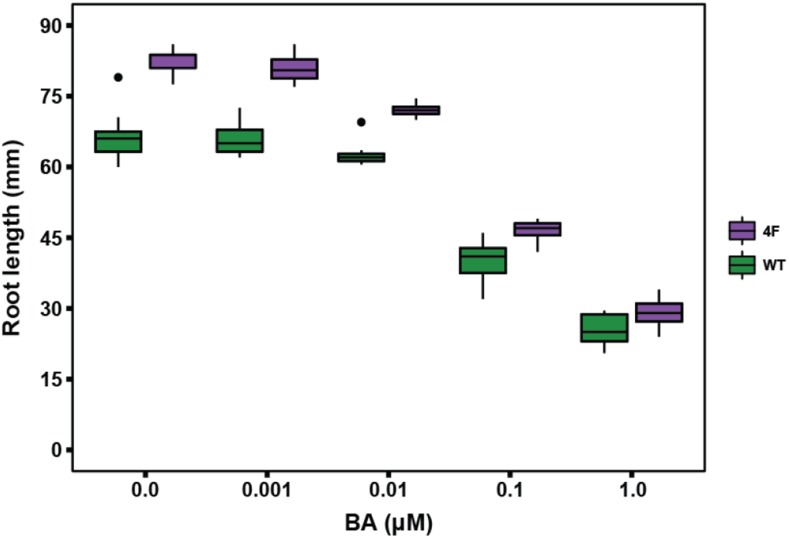
The data above suggests a model in which nitrate leads to elevated cytokinin biosynthesis, cytokinins activate the expression of target glutaredoxin genes, and glutaredoxins mediate the repression of primary root growth. Based on this model, we would expect that glutaredoxin-silenced lines would show decreased sensitivity to cytokinin-mediated inhibition of root growth. However, this does not appear to be the case. As shown in Figure 6, increasing levels of exogenous cytokinin consistently reduced primary root length in both the wild type and the glutaredoxin-silenced line 4F, though the roots of 4F remained longer than the wild type at all cytokinin concentrations. This result suggests that an alternative pathway independent of AtGRXS3/4/5/8 may exist to link cytokinins with primary root growth. It is possible that this alternative pathway could involve the other nitrate- and cytokinin-regulated glutaredoxins not targeted by our RNA interference construct (AtGRXS6 and AtGRXC11), given that functional redundancy is a common feature of many class III glutaredoxins (Xing and Zachgo, 2008; Ströher and Millar, 2012). Collectively, the results described above are consistent with the signaling pathway proposed in Figure 7, in which (1) nitrate activates cytokinin biosynthesis and root-to-shoot transport, (2) cytokinins up-regulate expression of the AtGRXS3,4,5,8 genes via the action of ARR1/10/12, and (3) the corresponding glutaredoxin enzymes act to inhibit primary root growth (Takei et al., 2004; Brenner et al., 2005; Ruzicka et al., 2009).
Figure 6.
Effect of exogenous cytokinins on primary root growth of the glutaredoxin-silenced line 4F and the wild type (WT). Analyses were performed on 11-d-old seedlings grown on vertical plates containing the noted concentrations of 6-benzylaminopurine (BA). The plots present the distribution of measurements for ≥ 6 replicates per experiment. Boxes depict the inter quartile range (IQR) that includes 50% of data points, horizontal lines represent the median value, vertical lines represent the distance from the hinge to highest/lowest value within the range of 1.5 × IQR, and data points beyond the 1.5 × IQR limit were considered outliers (dots).
Figure 7.
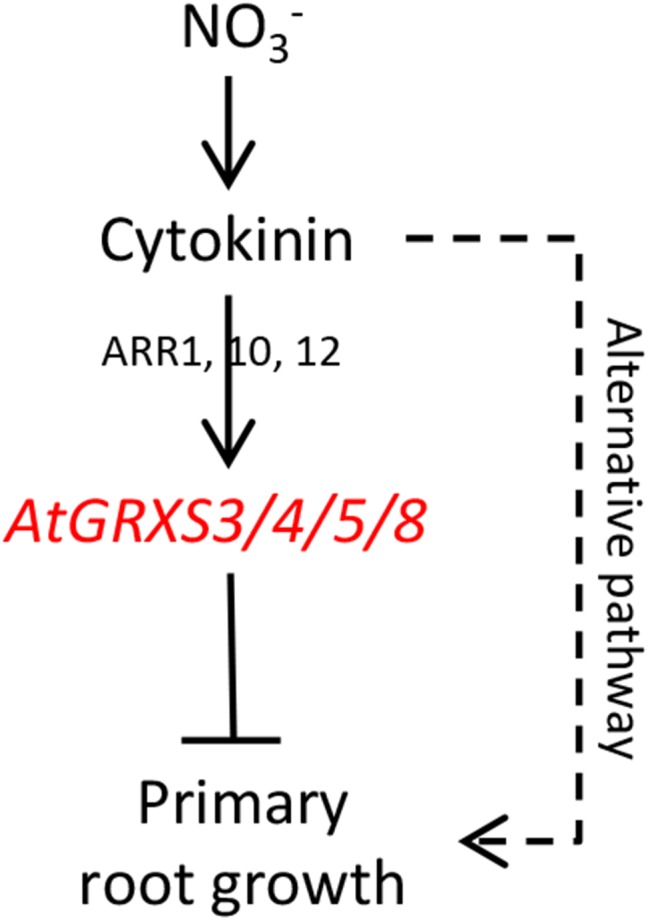
Proposed signal transduction pathway linking nitrate, cytokinin, and AtGRXS3/4/5/8 in the regulation of primary root growth.
DISCUSSION AND CONCLUSION
Although our initial experimental objective was simply to characterize the differential effects of nitrate and ammonium on the Arabidopsis shoot transcriptome, subsequent functional analyses of a group of nitrate-induced class III glutaredoxins revealed that these genes play a fundamental role in the regulation of primary root growth. The glutaredoxin gene family is greatly expanded in higher plants, with 31 glutaredoxin genes in Arabidopsis, compared to four in Escherichia coli and three in humans (Meyer et al., 2009). In all living systems, glutaredoxins are key players in maintaining cellular redox homeostasis and redox signaling. Biochemically, glutaredoxins function primarily in the reduction of protein-glutathione mixed disulfide bonds (deglutathionylation), often altering the activity of their target proteins (Ströher and Millar, 2012). Following a deglutathionylation reaction, most glutaredoxins are nonenzymatically reduced by glutathione, thus tying their activity to cellular redox poise. Although the link between glutaredoxins and primary root development in plants is novel, several previous studies have revealed connections between root redox status and growth. Suppressing glutathione biosynthesis in planta through the use of biosynthetic mutants or chemical inhibitors results in the repression of root growth, with primary root length directly correlated with glutathione pool size (Vernoux et al., 2000; Bashandy et al., 2010; Koprivova et al., 2010). The redox poise of the glutathione pool also appears critical, since a glutathione reductase2 mutant with a highly oxidized glutathione pool shows strongly decreased primary root growth (Yu et al., 2013). Strikingly, a very recent transcriptome analysis of the root meristemless1 glutathione biosynthesis mutant demonstrated that AtGRXS1, AtGRXS3, AtGRXS5, and AtGRXS6 (nitrate-regulated glutaredoxins) were all significantly down-regulated in the mutant line (Schnaubelt et al., 2015). Any connection between nitrate-regulated glutaredoxins and the glutathione pool should be considered tentative, however, given that at least some glutaredoxins are not reduced by glutathione (Zaffagnini et al., 2008) and none of the nitrate-regulated glutaredoxins has been purified and enzymatically characterized. Still, it appears that nitrate-regulated glutaredoxins, glutathione, and overall root redox poise may collectively influence root system development, though the specific mechanistic details of this network remain unclear.
While class I and II glutaredoxins are typically associated with oxidative stress response and the assembly of iron sulfur clusters, only a handful of the plant-specific class III glutaredoxins have been studied previously. Of these, the best characterized are AtGRXC7 (ROXY1) and AtGRXC8 (ROXY2), which are involved in stamen and petal development, and AtGRXC9 (ROXY19), which is involved in plant defense (Xing et al., 2005; Ndamukong et al., 2007; Xing and Zachgo, 2008). The activity of these three glutaredoxins is associated with their ability to bind to (and presumably posttranslationally regulate) TGA-type transcription factors. A recent analysis by Zander et al. (2012) demonstrated that all tested class III glutaredoxins, including AtGRXS5, are capable of binding to the TGA2 transcription factor in a yeast two-hybrid assay (Zander et al., 2012). Though subcellular localization has been characterized for only a few class III glutaredoxins (AtGRXC7, -13, -14; AtGRXS9, -11), all of these display nuclear-cytosolic localization (Li et al., 2009; Li et al., 2011), suggesting that protein interactions with TGA transcription factors are biologically feasible. Although the subcellular localization of the AtGRXS3/4/5/8 proteins is unknown, they all contain the same C-terminal −ALWL domain that appears to be required for the glutaredoxin-TGA interaction (Zander et al., 2012). Interestingly, a recent study has implicated the TGA1 and TGA4 transcription factors as important regulators of nitrate response in Arabidopsis (Alvarez et al., 2014). Clearly, further study is merited to determine whether regulation of primary root growth by AtGRXS3/4/5/8 is linked to the binding and posttranslational modification of TGA transcription factors in vivo.
Compared to our relatively sophisticated understanding of the effects of nitrogen on the initiation and elongation of lateral roots, the regulation of primary root growth by nitrogen is rather poorly understood (Forde, 2014; Giehl and von Wirén, 2014). For example, nitrate foraging, the precise regulation of root system development to maximize utilization of nitrate-rich areas of the soil, has been studied primarily in relation to lateral root development (Zhang and Forde, 1998; Remans et al., 2006; Guan et al., 2014). The seminal work of Zhang and Forde (1998) showed that the MADS-box transcription factor ANR1 regulates the nitrate-dependent outgrowth of lateral roots, allowing plants to maximize “horizontal coverage” of nitrate-rich soil patches (Zhang and Forde, 1998). The model proposed in Figure 3B suggests that nitrate-regulated glutaredoxins may play a complementary role in nitrate foraging, allowing plants to slow the growth of the primary root in nitrate-rich regions at the same time as they are maximizing lateral root growth. Since glutaredoxins (and cytokinin biosynthesis) are not up-regulated by ammonium, this response would allow Arabidopsis to maximize utilization of its preferred nitrogen source, nitrate, while minimizing potential ammonium toxicity. The question of whether nitrate has a positive, negative, or neutral effect on primary root growth in Arabidopsis has produced a variety of apparently conflicting answers (Vidal et al., 2010; Cerutti and Delatorre, 2013; Forde, 2014), and may significantly depend on the plant growth conditions, nitrate concentrations, and ecotypes utilized. Our model aligns with the work of Tian et al. (2005), who showed that nitrate increases cytokinin levels and decreases primary root growth in maize, and the work of Vidal et al. (2010), who demonstrated that nitrate inhibits primary root growth in Arabidopsis via a regulatory module involving the microRNA miR393 and the auxin receptor protein AFB3 (Tian et al., 2005; Vidal et al., 2010). Additional studies will be needed to determine whether glutaredoxin-mediated regulation of primary root growth intersects with this miR393/AFB3 pathway in Arabidopsis.
The signal transduction pathway summarized in Figure 7 integrates current models of primary root development (Ruzicka et al., 2009; Ruffel et al., 2011) with our functional analyses of nitrate-induced glutaredoxins. The cytokinin biosynthesis gene AtIPT3 is specifically up-regulated by nitrate in Arabidopsis, leading to elevated cytokinin levels in both roots and shoots (Kiba et al., 2005). Ruffel et al. (2011) proposed that cytokinin acts as a systemic signal underlying a root-shoot-root signaling relay that communicates plant nitrate status (Ruffel et al., 2011). In this respect, it is interesting that both IPT3 (Takei et al., 2004) and AtGRXS3/4/5/8 (Zhao et al., 2005; Brady et al., 2007; Ruiz-Medrano et al., 2011) appear to be expressed primarily in phloem, which is the tissue involved in transport of isopentenyladenine-type and trans-zeatin-type cytokinins (Hirose et al., 2008). Both our data and several previous global expression profiling studies clearly demonstrate that AtGRXS3, -4, -5, and -8 are strongly up-regulated by cytokinins. Most recently, two meta-analyses of microarray and RNA-seq data placed AtGRXS4 and AtGRXS8 among a group of consistently regulated “marker genes” for the cytokinin-mediated transcriptional response (Brenner et al., 2012; Bhargava et al., 2013). Brenner et al. (2012) hypothesized that these glutaredoxins might play a role in a putative oxidative stress response activated by cytokinins. However, our results suggest that AtGRXS3/4/5/8 are instead integrating information about plant nitrate status and cytokinin signaling to regulate the growth of the Arabidopsis primary root. A very recent publication further supports the connections between glutaredoxins and nitrate status. The class III glutaredoxin OsGRX6 (also known as OsGRXS5) was identified in a screen for genes involved in nitrogen use efficiency in rice (Oryza sativa). Ectopic overexpression of this gene in rice resulted in an array of developmental phenotypes, increased gibberellin and cytokinin levels, and an increase in total nitrogen content in shoots and seeds (El-Kereamy et al., 2015). Although OsGRX6 is rather distantly related to AtGRXS3/4/5/8 phylogenetically (Ströher and Millar, 2012), it is clear that class III glutaredoxins may play important roles in root hormone biology and mineral nutrition.
MATERIALS AND METHODS
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (wild type), the cytokinin oxidase overexpressing line CKX3-9 (Werner et al., 2003) in the Columbia-0 background, the ARR1/10/12 mutant in the Columbia-0 background, an AtGRXS3 transposon mutant line in the Nössen background (RATM12-4682-1_G), and two parental lines used to create the AtGRXS3 mutant (CS8514 and CS8538) were utilized in these studies. To create glutaredoxin-silenced transgenic plants, the AtGRXS3 gene (nucleotides 3–309 of the coding sequence) was PCR amplified using the Glut700-genF and Glut700-genR primers (Supplemental Table S3) and the Verbatim High Fidelity Polymerase (Thermo Scientific). The PCR product was cloned into pCRBluntII-TOPO (Life Technologies) and sequence verified. Using two double digests (XhoI/KpnI; XbaI/HindIII) and subsequent ligations, the AtGRXS3 sequence was cloned into the pKannibal RNA interference vector (Wesley et al., 2001). The glutaredoxin-silencing construct was excised from pKannibal using a NotI digest, and was ligated into the pDU99.2215 binary vector (Escobar et al., 2001), generating pESil700. This vector was electroporated into Agrobacterium tumefaciens EHA101, and Arabidopsis Columbia-0 plants were subsequently transformed using the floral dip method (Clough and Bent, 1998). Transformants were selected on plates of half-strength Murashige and Skoog medium (pH 6.0) containing 50 mg/L kanamycin.
Hydroponic growth of Arabidopsis was performed as previously described (Patterson et al., 2010), with plants at growth stage 5.10 (Boyes et al., 2001) transferred to nitrogen-free media for 26 h, followed by transfer to media containing 1 mm KNO3, 0.5 mm (NH4)SO4, or 0.5 mm K2SO4. For cytokinin treatment, hydroponically grown plants were nitrogen starved for 26 h and then the leaves were sprayed with a solution of 20 µm trans-zeatin dissolved in 0.02% (v/v) dimethyl sulfoxide. For root growth assays, seeds were sown on plates containing half-strength Murashige and Skoog medium, 1% Suc, pH 6.0. Plates were incubated at 4°C for 2 d and then transferred to a growth chamber (16 h light period with approximately 100 µmol m−2 s−1 illumination at 22°C, 8 h dark period at 18°C) for 5 d. Seedlings were then transferred to the surface of vertically oriented plates containing half-strength Somerville and Ogren medium (Somerville and Ogren, 1982) with the reported micronutrient mix at 1× concentration. These plates do not contain an exogenous carbon source, and nitrate (4.5 mm) is the only nitrogen source. The plates were incubated under the same growth conditions and root length was measured after 5 to 6 d. Hydroponic growth of plants on ammonium succinate medium was carried out essentially as described by Vidal et al. (2010) with some modifications. Seeds were sown on a 250 µm stainless steel mesh cut to fit snugly in a GA-7 tissue culture vessel (Magenta). The vessel was filled with Murashige and Skoog basal salt medium without nitrogen (Phytotechnology Laboratories) supplemented with 0.5 mm ammonium succinate and 3 mm Suc. Vessels were incubated at 4°C for 3 d and then transferred to a growth chamber under the conditions described above. After 9 d, 5 mm KNO3 or 5 mm KCl was added to the vessels. Primary root length was assayed 3 d after KNO3/KCl treatment.
RNA isolation was performed using an RNeasy Plant Kit (Qiagen), with the RNA subsequently DNAse-treated using a Turbo DNA-free kit (Ambion). RNA quality and concentration were determined spectrophotometrically and RNA integrity was assayed by formaldehyde agarose gel electrophoresis (Patterson et al., 2010). cDNA synthesis was performed using a Verso cDNA synthesis kit (Thermo Scientific) with 500 to 1,000 ng total RNA and oligo(dT) primers. Real-time PCR was carried out as previously described using a Rotor Gene 6000 Real-Time Cycler (Corbett Research; Patterson et al., 2010). Real-time PCR data were normalized using the constitutively expressed standard gene At5g37510, which encodes the 76 kD subunit of respiratory complex I (Svensson and Rasmusson, 2001). Primers utilized in real-time PCR are included in Supplemental Table S3.
The synthesis of biotin-labeled aRNA, hybridization of aRNA to the Affymetrix ATH1 GeneChip, scanning, and microarray data analysis using RMA (Bolstad et al., 2003) were all performed as previously described (Patterson et al., 2010). Methods associated with supplemental figures are included in the supplemental figure files.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Organ specificity in transcriptional responses to nitrate and ammonium.
Supplemental Figure S2. Short-term ammonium and nitrate treatment do not induce oxidative stress in Arabidopsis shoots.
Supplemental Figure S3. Regulation of glutaredoxin gene expression in Arabidopsis roots.
Supplemental Figure S4. Sequence and regulatory conservation of AtGRXS3, AtGRXS4, AtGRXS5, and AtGRXS8.
Supplemental Figure S5. Nitrate-mediated inhibition of root growth in the AtGRXS3 mutant and the 35S:CKX3 transgenic line.
Supplemental Figure S6. Comparison of global transcriptional responses to nitrate and cytokinin.
Supplemental Figure S7. Increase in the number of root meristem cortical cells in AtGRX3 RNA interference lines (1A and 4F).
Supplemental Table S1. List of genes specifically regulated by nitrate in Arabidopsis shoots.
Supplemental Table S2. List of genes specifically regulated by ammonium in Arabidopsis shoots.
Supplemental Table S3. Primers utilized in PCR and real-time PCR.
Supplementary Material
Acknowledgments
We thank Thomas Schmülling for providing the 35S:CKX3 seeds and Takafumi Yamashino for providing the ARR1/10/12 mutant seeds. We are also grateful to Elena Vidal for advice on the preparation of ammonium succinate growth medium. Microarray hybridizations and initial bioinformatics analyses of array data were performed by The Scripps Research Institute DNA Array Core Facility.
Glossary
- RT-PCR
reverse transcription PCR
Footnotes
Articles can be viewed without a subscription.
References
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-López O, Tamayo KP, Aceituno F, Gómez I, Ruffel S, Lejay L, et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J 80: 1–13 [DOI] [PubMed] [Google Scholar]
- Argyros RD, Mathews DE, Chiang YH, Palmer CM, Thibault DM, Etheridge N, Argyros DA, Mason MG, Kieber JJ, Schaller GE (2008) Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20: 2102–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C, Bashandy T, Cela J, Delorme-Hinoux V, Riondet C, Reichheld JP (2015) A comprehensive study of thiol reduction gene expression under stress conditions in Arabidopsis thaliana. Plant Cell Environ 38: 299–314 [DOI] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Boudsocq S, Niboyet A, Lata JC, Raynaud X, Loeuille N, Mathieu J, Blouin M, Abbadie L, Barot S (2012) Plant preference for ammonium versus nitrate: a neglected determinant of ecosystem functioning? Am Nat 180: 60–69 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T (2012) Gene regulation by cytokinin in Arabidopsis. Front Plant Sci 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ (2002) NH4+ toxicity in higher plants: a critical review. J Plant Physiol 159: 567–584 [Google Scholar]
- Cerutti T, Delatorre CA (2013) Nitrogen and phosphorus interaction and cytokinin: responses of the primary root of Arabidopsis thaliana and the pdr1 mutant. Plant Sci 198: 91–97 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S (2007) Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol 17: 678–682 [DOI] [PubMed] [Google Scholar]
- El-Kereamy A, Bi YM, Mahmood K, Ranathunge K, Yaish MW, Nambara E, Rothstein SJ (2015) Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front Plant Sci 6: 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM (2001) RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc Natl Acad Sci USA 98: 13437–13442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG. (2014) Nitrogen signalling pathways shaping root system architecture: an update. Curr Opin Plant Biol 21: 30–36 [DOI] [PubMed] [Google Scholar]
- Giehl RF, von Wirén N (2014) Root nutrient foraging. Plant Physiol 166: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber N, Galloway JN (2008) An Earth-system perspective of the global nitrogen cycle. Nature 451: 293–296 [DOI] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM (2014) Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc Natl Acad Sci USA 111: 15267–15272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59: 75–83 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B (2012) Cytokinin signaling networks. Annu Rev Plant Biol 63: 353–380 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T (2008) Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol 49: 47–57 [DOI] [PubMed] [Google Scholar]
- Ito T, Motohashi R, Kuromori T, Noutoshi Y, Seki M, Kamiya A, Mizukado S, Sakurai T, Shinozaki K (2005) A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol 46: 1149–1153 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T (2005) Combinatorial microarray analysis revealing arabidopsis genes implicated in cytokinin responses through the His->Asp phosphorelay circuitry. Plant Cell Physiol 46: 339–355 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Mugford ST, Kopriva S (2010) Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- La Camera S, L’haridon F, Astier J, Zander M, Abou-Mansour E, Page G, Thurow C, Wendehenne D, Gatz C, Métraux JP, et al. (2011) The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J 68: 507–519 [DOI] [PubMed] [Google Scholar]
- Laporte D, Olate E, Salinas P, Salazar M, Jordana X, Holuigue L (2012) Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J Exp Bot 63: 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li G, Kronzucker HJ, Baluška F, Shi W (2014) Ammonium stress in Arabidopsis: signaling, genetic loci, and physiological targets. Trends Plant Sci 19: 107–114 [DOI] [PubMed] [Google Scholar]
- Li P, Ham BK, Lucas WJ (2011) CmRBP50 protein phosphorylation is essential for assembly of a stable phloem-mobile high-affinity ribonucleoprotein complex. J Biol Chem 286: 23142–23149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S (2009) Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP (2009) Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Møller AL, Pedas P, Andersen B, Svensson B, Schjoerring JK, Finnie C (2011) Responses of barley root and shoot proteomes to long-term nitrogen deficiency, short-term nitrogen starvation and ammonium. Plant Cell Environ 34: 2024–2037 [DOI] [PubMed] [Google Scholar]
- Mutwil M, Obro J, Willats WG, Persson S (2008) GeneCAT--novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Patterson K, Cakmak T, Cooper A, Lager I, Rasmusson AG, Escobar MA (2010) Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ 33: 1486–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmülling T (2006) Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM (2011) Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA 108: 18524–18529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano R, Xoconostle-Cázares B, Ham BK, Li G, Lucas WJ (2011) Vascular expression in Arabidopsis is predicted by the frequency of CT/GA-rich repeats in gene promoters. Plant J 67: 130–144 [DOI] [PubMed] [Google Scholar]
- Ruzicka K, Simásková M, Duclercq J, Petrásek J, Zazímalová E, Simon S, Friml J, Van Montagu MC, Benková E (2009) Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc Natl Acad Sci USA 106: 4284–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Sarasketa A, González-Moro MB, González-Murua C, Marino D (2014) Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J Exp Bot 65: 6023–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schnaubelt D, Queval G, Dong Y, Diaz-Vivancos P, Makgopa ME, Howell G, De Simone A, Bai J, Hannah MA, Foyer CH (2015) Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ 38: 266–279 [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Isolation of Photorespiration Mutants in Arabidopsis thaliana. In Edelman M, Hallick RB, Chua N-H, eds, Methods in Chloroplast Biology. Elsevier Biomedical Press, New York, pp 129–138 [Google Scholar]
- Ströher E, Millar AH (2012) The biological roles of glutaredoxins. Biochem J 446: 333–348 [DOI] [PubMed] [Google Scholar]
- Svensson AS, Rasmusson AG (2001) Light-dependent gene expression for proteins in the respiratory chain of potato leaves. Plant J 28: 73–82 [DOI] [PubMed] [Google Scholar]
- Takei K, Ueda N, Aoki K, Kuromori T, Hirayama T, Shinozaki K, Yamaya T, Sakakibara H (2004) AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol 45: 1053–1062 [DOI] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277 [DOI] [PubMed] [Google Scholar]
- Tian QY, Chen FJ, Zhang FS, Mi GH (2005) Possible involvement of cytokinin in nitrate-mediated root growth in maize. Plant Soil 277: 185–196 [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Filleur S, Gan Y, Forde BG (2005) Signaling mechanisms integrating root and shoot responses to changes in the nitrogen supply. Photosynth Res 83: 239–250 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S (2005) ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Yu X, Pasternak T, Eiblmeier M, Ditengou F, Kochersperger P, Sun J, Wang H, Rennenberg H, Teale W, Paponov I, et al. (2013) Plastid-localized glutathione reductase2-regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 25: 4451–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD (2008) Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J Biol Chem 283: 8868–8876 [DOI] [PubMed] [Google Scholar]
- Zander M, Chen S, Imkampe J, Thurow C, Gatz C (2012) Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol Plant 5: 831–840 [DOI] [PubMed] [Google Scholar]
- Zhang H, Forde BG (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.