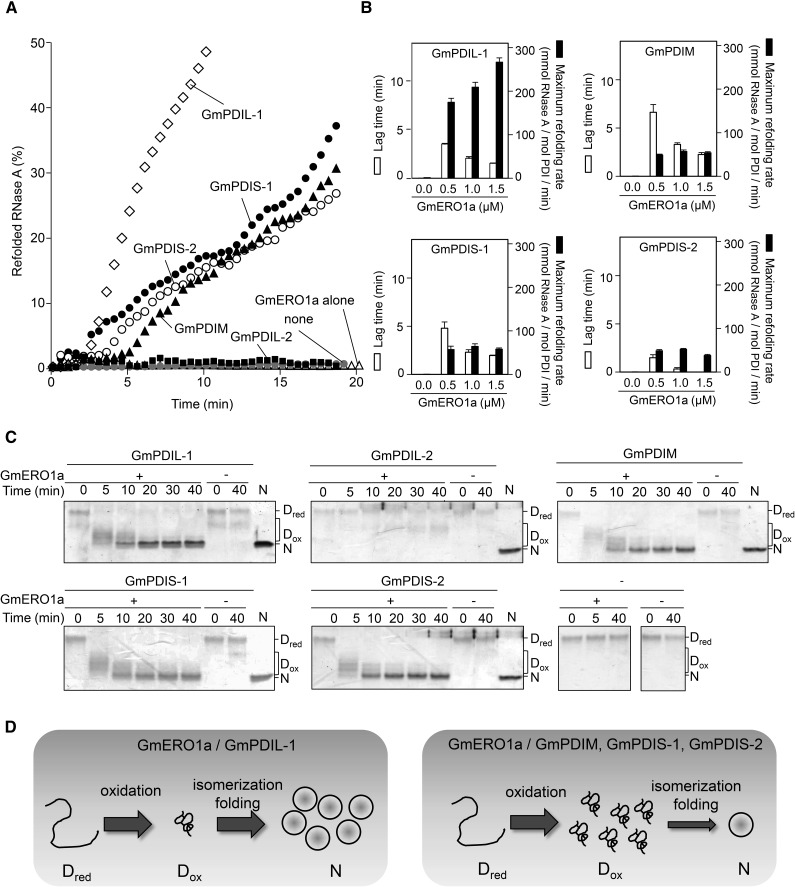
Figure 4.
Reconstitution of oxidative protein folding with ER oxidoreductases and GmERO1a in vitro. A, GmERO1a (1 μm) without (GmERO1a alone), or with 3 μm of each ER oxidoreductase, was incubated with reduced and denatured RNase A (8 μm), and the recovered RNase A activity was assayed. “None” shows refolding of RNase A alone. B, Lag time (white bars) and maximum refolding rate (black bars) of reduced and denatured RNase A (8 μm) by 3 μm each ER oxidoreductase in the presence of GmERO1a. Data are represented as mean ± se of n = 3. C, Refolding of reduced and denatured RNase A in the absence (−) or presence (+) of GmERO1a and each ER oxidoreductase, quenched with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid, and was analyzed by nonreducing SDS-PAGE. Dred, reduced and denatured RNase A; Dox, denatured RNase A with nonnative disulfides; N, native RNase A. D, Model of refolding of RNase A by ER oxidoreductases in the presence of GmERO1a. Left, GmPDIL-1 transfers nonnative disulfide bonds to Dred. Dox is promptly folded into the native form accompanied by the isomerization of disulfide bonds. Right, GmPDIM, GmPDIS-1, or GmPDIS-2 transfers nonnative disulfide bonds to Dred. Dox accumulates due to rate-limiting isomerization activities of GmPDIM, GmPDIS-1, and GmPDIS-2.