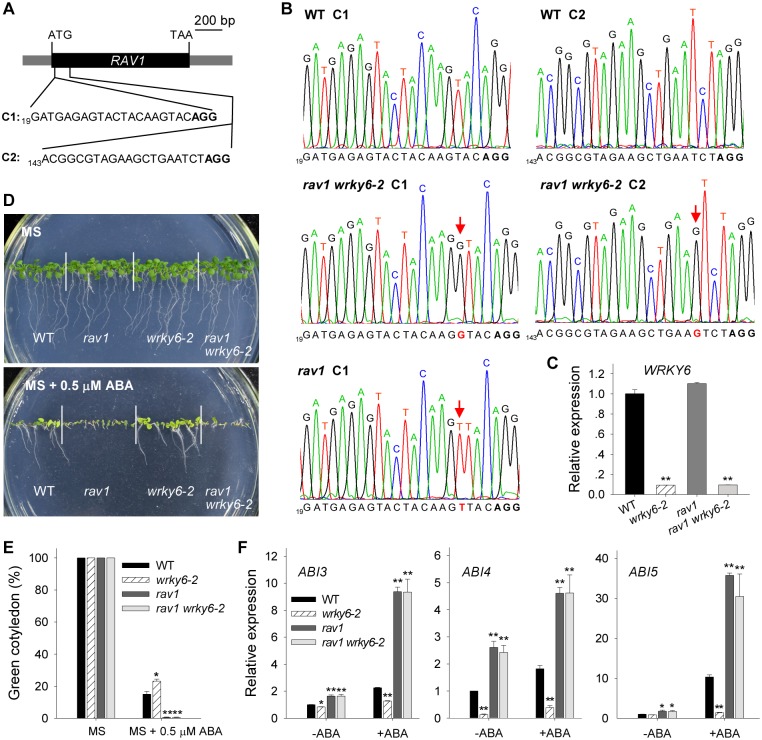
Fig 10. Disruption of RAV1 abolishes the ABA-insensitivity of the wrky6 mutant.
A, Diagram of RAV1 showing two target sites (C1 and C2) for CRISPR/Cas9 technology. PAM motifs are marked with bold letters. The coding sequence (CDS) and untranslated regions (UTR) of RAV1 are indicated by black box and gray boxes, separately. B, The rav1 mutant and rav1 wrky6-2 double mutant were generated by CRISPR/Cas9 technology. The mutation in the RAV1 gene is evaluated by sequencing, and the mutant sites in RAV1 are indicated by red letters and arrows. C, qRT-PCR analysis of WRKY6 expression in the wrky6-2 mutant, rav1 mutant, rav1 wrky6-2 double mutant and wild-type plants (WT). Data are shown as mean ± SE (n = 3). D, Phenotypic comparison. Imbibed seeds were transferred to MS or MS + 0.5 μM ABA medium for 10 d. E, Cotyledon-greening analysis. Imbibed seeds were transferred to MS or MS + 0.5 μM ABA medium for 7 d before determining cotyledon-greening percentage. Data are shown as mean ± SE (n = 3). F, qRT-PCR analysis of ABIs in the wrky6-2 mutant, rav1 mutant, rav1 wrky6-2 double mutant and wild-type plants (WT) treated with or without exogenous ABA treatment. The 7-d-old seedlings were transferred to MS solution with or without 100 μM ABA for 3 h, and then the seedlings were harvested for qRT-PCR. Data are shown as mean ± SE (n = 3). Asterisks indicate statistically significant differences compared with relevant wild-type plants (WT): *, P 0.05; **, P 0.01.