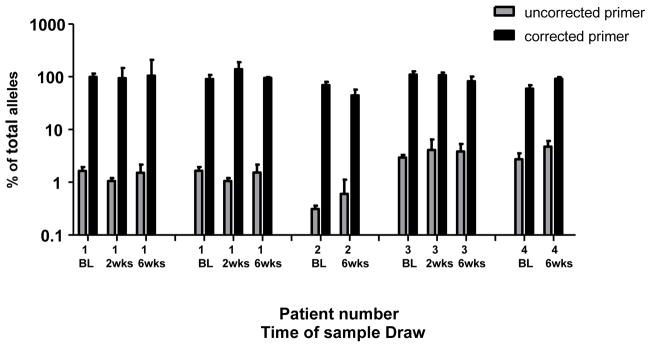
Figure 1.
Comparison of allele-specific PCR efficiency of patient samples between primers based on HIV-1 subtype C consensus sequences and primers based on patient-specific HIV-1 consensus sequences at codon 103. The total amplification efficiency of patient samples 1–4 (Table 1) was calculated as the sum of the efficiencies of detection of the wild-type and mutant alleles. Shown are results using the uncorrected consensus primers (grey) and the corrected primers (black) with error bars representing standard deviations of triplicate observations for samples taken immediately before single-dose nevirapine (BL) and 2 and/or 6 weeks following treatment. Patient 1 has 2 data sets; the 1st are samples amplified with primers that match the HIV-1 sequence infecting patient 1, the 2nd are samples amplified with primers only corrected to match the sequence at codon 104 in patient 1.