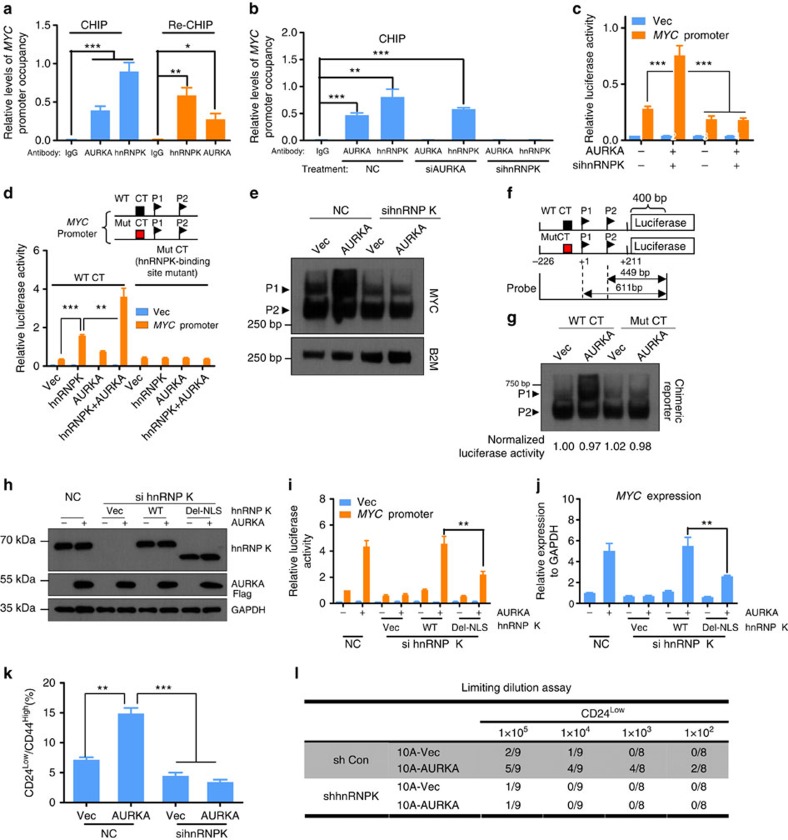
Figure 4. hnRNP K is required for AURKA to activate MYC transcription and enhance BCSC phenotype.
(a) Chromatin from MDA-MB-231 cells was extracted for ChIP and re-ChIP analysis. Results were normalized by input. (b) MDA-MB-231 cells were transfected with siRNA against negative control (NC), AURKA or hnRNP K for 48 h. ChIP assays were performed. (c) MDA-MB-231 cells overexpressing HA-tagged AURKA were transfected with hnRNP K or NC siRNA. After 24 h, cells were transfected with MYC promoter reporter (MYC) or basic reporter (Vec) along with pRL-TK for another 24 h. Dual-luciferase reporter assay was performed. (d) Mutated reporter along with pRL-TK were co-transfected with AURKA or/and hnRNP K plasmids for 24 h. Dual-luciferase reporter assay was performed. (e) 293T cells overexpressing HA-AURKA were transfected with hnRNP K or NC siRNA for 48 h. S1 nuclease protection assay (SNPA) was performed to monitor MYC P1 and P2 transcripts. (f) Structure of chimeric gene consists of firefly luciferase and MYC promoter (−226/+211) (upper panel). Lower panel shows probes used for SNPA. ‘CT' represented CT element. The mutant chimeric gene was mutated at hnRNP K-binding site. (g) Chimeric gene or its mutant were co-transfected with AURKA and pRL-TK plasmids into 293T cells for 24 h. SNPA was performed using the probes shown in f. In parallel, a fraction of these cells was evaluated for Renilla luciferase activity, which reflects the transfection efficiency. (h,i,j) 293T cells were transfected with hnRNP K siRNA for 24 h. Cells were then co-transfected with WT or NLS-deleted hnRNP K, AURKA and MYC promoter or basic reporter along with pRL-TK for another 24 h. Transfected cells were harvested for immunoblotting (IB) (h), dual-luciferase reporter (i) and real-time PCR (j) analysis. (k) MCF-10A cells overexpressing HA-AURKA were transfected with hnRNP K or NC siRNA. After 48 h, cells were collected to analyse CD24/CD44. (l) hnRNP K was knocked down in AURKA-overexpressing or control 10A-K-Ras (G12V) cells. Cells were sorted according to CD24 expression. CD24Low population was used to perform limiting dilution assays. Bar represented the means±s.e.m. of three independent experiments (analysis of variance (ANOVA) followed by least significant difference (LSD) test; *P<0.05, **P<0.01, ***P<0.001).