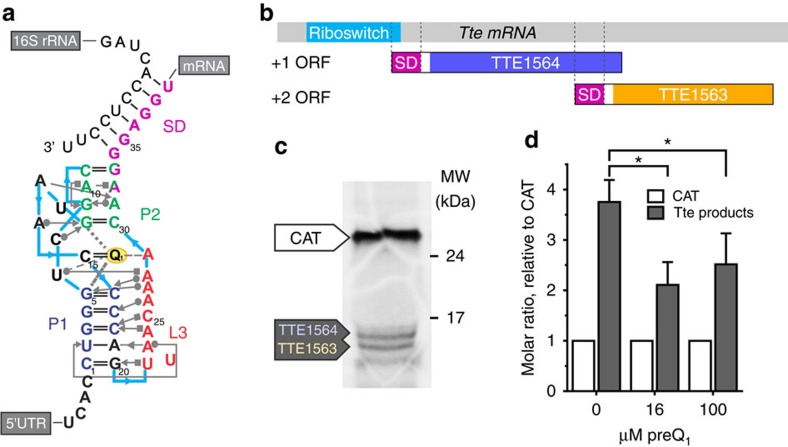
Figure 1. In vitro translation of Tte mRNA.
(a) Structural map of the Tte preQ1 translational riboswitch displayed with Leontis–Westhof notations57. The SD sequence (purple) partially overlaps the P2 stem nucleotides (green). Formation of the P2 stem interferes with proper base pairing between the anti-SD sequence at the 3′ end of the 16S rRNA and the SD sequence of the Tte mRNA. (b) Schematic of Tte mRNA used for in vitro translation assays. The putative mRNA transcript is bicistronic, containing the overlapping reading frames for TTE1564 and TTE1563. The preQ1 riboswitch aptamer (light blue) overlaps with a portion of the SD sequence (purple) of TTE1564. (c) Example autoradiograph of in vitro translation products. Tte mRNA was translated using L-[35S]-Cys in the presence of the control mRNA encoding CAT at a 4:1 ratio of Tte:CAT mRNA. Molecular weight markers are indicated on the right (full-length gel is shown in Supplementary Fig. 11). (d) Quantification of in vitro translation products as a function of preQ1 concentration. The total protein produced from the Tte mRNA (sum of TTE1564 and TTE1563 bands, grey bar) is reported relative to the intensity of the CAT product in the same lane, after normalizing for the cysteine content of each protein (5, 1 and 1 for CAT, TTE1564 and TTE1563, respectively). The results represent the mean±s.d. of three replicates (*P<0.05).