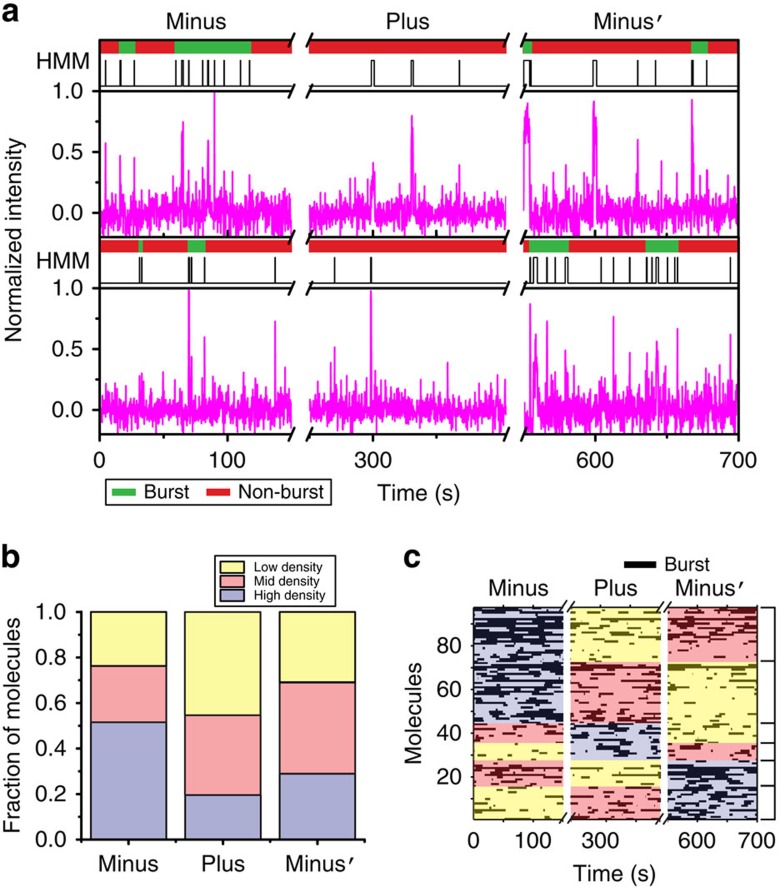
Figure 5. Single mRNA molecules can undergo conformational switching depending on their environment.
(a) Exemplary single-molecule trajectories from ligand-jump SiM-KARTS experiments composed of three time segments. Anti-SD probe binding to the same set of individual Tte mRNA molecules is monitored first in the absence of preQ1 (Minus), then in the presence of 16 μM preQ1 (Plus) and again in the absence of preQ1 (Minus′). Each axis break represents a 50-s dark period between segments during which buffer was exchanged. (b) Distribution of burst density rankings for each segment of an individual molecule's trajectory, for all molecules in the ligand-jump SiM-KARTS experiment (N=97). (c) Rastergram displaying the bursting behaviour of the Tte mRNA molecules in b. Bursts are displayed as black bars. Individual probe-binding events (spikes) are omitted for clarity.