Fig. 7.
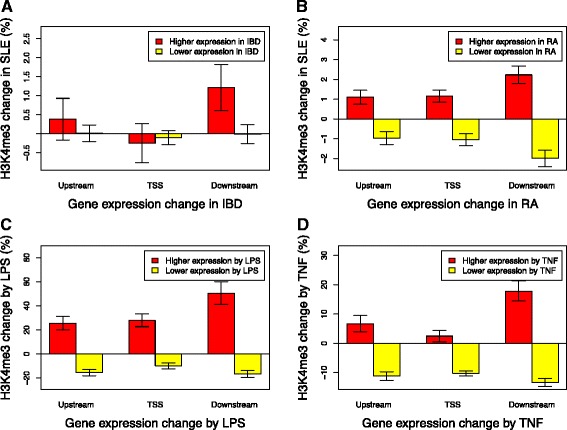
Validation in other inflammatory models. a Genes with significantly increased or decreased transcription in inflammatory bowel disease (IBD) were obtained from our unpublished RNA-seq data. The average H3K4me3 changes in our unmatched SLE samples were calculated at three different regions, showing that the only significant change was the increase of downstream H3K4me3 at TSS of genes with increased transcription. b The same analysis was applied to a published data set (GSE10500) comparing the transcriptome of rheumatoid arthritis (RA) patients and controls in macrophage. c This analysis used matching RNA-seq and H3K4me3 data obtained from primary monocytes treated with LPS or cell culture medium as control (GSE58310). Average H3K4me3 changes of gene sets with increased or decreased transcription by LPS were calculated for the same three regions. Error bars indicate that average H3K4me3 was changed significantly towards the same direction as the change of transcription. d The same analysis was applied to matching ChIP-seq H3K4me3 and microarray transcriptome data generated from TNF-treated endothelium (GSE54000). All plots normalized H3K4me3 data to make the average change of all genes equal to zero at each region
