Abstract
The significance of bacteria for eukaryotic functioning is increasingly recognized. Coral reef ecosystems critically rely on the relationship between coral hosts and their intracellular photosynthetic dinoflagellates, but the role of the associated bacteria remains largely theoretical. Here, we set out to relate coral‐associated bacterial communities of the fungid host species Ctenactis echinata to environmental settings (geographic location, substrate cover, summer/winter, nutrient and suspended matter concentrations) and coral host abundance. We show that bacterial diversity of C. echinata aligns with ecological differences between sites and that coral colonies sampled at the species’ preferred habitats are primarily structured by one bacterial taxon (genus Endozoicomonas) representing more than 60% of all bacteria. In contrast, host microbiomes from lower populated coral habitats are less structured and more diverse. Our study demonstrates that the content and structure of the coral microbiome aligns with environmental differences and denotes habitat adequacy. Availability of a range of coral host habitats might be important for the conservation of distinct microbiome structures and diversity.
Keywords: coral reef, ecological niche, holobiont, metaorganism, microbiome, symbiosis
Introduction
Recent advancements in sequencing technology have led to a new understanding of the role of micro‐organisms in shaping animal biology emphasizing the diversity and functional capacity of bacteria, and challenging our views on what constitutes a genome or an organism (McFall‐Ngai et al. 2013). While tropical shallow water corals have long been recognized to exist in close and obligate relationships with endosymbiotic unicellular algae (also referred to as zooxanthellae) of the genus Symbiodinium (Muscatine & Cernichiari 1969), the importance of the diverse community of bacteria became only recently established (Rosenberg et al. 2007). This functional metaorganism consisting of the coral animal host, its photosynthetic algal symbionts, and microbial assemblage is termed the coral holobiont (Rosenberg et al. 2007). Coral‐associated bacteria are shown to confer immunity (Kelman et al. 2006) and to support the host's metabolic demands (Lesser et al. 2004). They are rich in abundance as well as in diversity (Rohwer et al. 2002) and differ between holobiont compartments such as tissue, mucus or skeleton (Li et al. 2014). Their diversity furthermore differs from that of assemblages present in the surrounding water column (Frias‐Lopez et al. 2002; Roder et al. 2014) and the prevailing bacterial community is host species specific (Rohwer et al. 2002; Sunagawa et al. 2010). And even though intracolonial variation has been documented (Li et al. 2013), bacterial assemblages associated with corals are well structured with distinct operational taxonomic units (OTU) frequently being highly abundant (Rohwer et al. 2001; Bayer et al. 2013a,b). Differences in microbial communities across coral species are assumed to be due to different corals associating with different microbes of similar function rather than phylogenetic affiliation (Li et al. 2013; Kelly et al. 2014). Nevertheless, changes in response to season (Littman et al. 2009) or geographic location (Koren & Rosenberg 2006) have been documented, and coral‐associated bacterial communities are sensitive towards environmental insult, experiencing large shifts during bleaching or disease (Bourne et al. 2008; Roder et al. 2014). It has been proposed that dynamics in coral‐associated microbial populations are an important mechanism for the holobiont to rapidly acclimate to changes in the environment (Reshef et al. 2006), but to which degree a coral's microbiome is structured by environmental conditions, temporal factors, or host phylogeny and physiology remains elusive and, to date, a comprehensive approach analysing coral microbiota dynamics in space and time is lacking.
Here, we set out to explore the variability of bacteria associated with the fungid coral Ctenactis echinata during summer and winter and across four habitats in the central Red Sea to further understand how environmental conditions and the coral microbiome structure relate. To do this, we ecologically described fore‐ and back‐reef environments of nearshore and offshore coral reefs detailing substrate condition, water temperature, nutrients, suspended matter concentrations and abundance of C. echinata and we compared these data to the associated bacterial communities of C. echinata across all sites.
Materials and methods
Sampling
Between 1 and 5 whole unattached and visually healthy polyps of Ctenactis echinata of equivalent size classes (<10 cm length) were collected across four reefs and their respective fore‐ and back‐reef environments over two sampling dates at 4–7 m depth using SCUBA in the central Red Sea. Each of two reefs denoted nearshore (i.e. Inner Fsar and Al Quad) and offshore (i.e. Abu Roma and Shib Nazaar) environments and were combined to provide between three and eight coral samples for any combination of fore‐reef or back‐reef and nearshore or offshore environments (Fig. 1). Details on sampling location, transect and environmental data collection and bacterial community sampling are provided as supplementary information (Table S1, Supporting information). Sampling took place on two occasions, once during summer (August 2011) and once during winter (February 2012) along exposed fore‐reef and sheltered back‐reef sides of two offshore (>25 km distance to shore) and two nearshore (<5 km distance to shore) reefs (Table S1, Supporting information). Samples were handled by wearing gloves and immediately transferred into sterile Whirl‐Pak bags after collection. Upon arrival on board, samples were rinsed with filtered (0.22 μm) sea water to remove loosely associated microbes. Samples were subsequently wrapped in aluminium foil and flash frozen in liquid nitrogen until analysis. At each sampling site, a water sample was collected in sterile cubitainers (1 L) and kept on ice until further processing (see below). Temperature at the study sites during sampling was recorded using a conventional thermometer.
Figure 1.
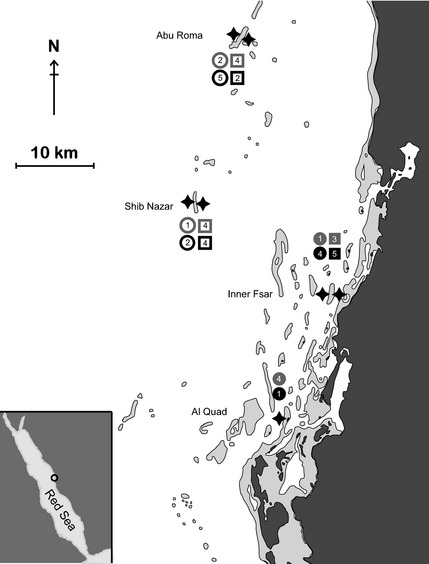
Map of study sites. Offshore (Abu Roma and Shib Nazar) and nearshore (Al Quad and Inner Fsar) coral reef sites were sampled from exposed (i.e. ocean facing) and sheltered (i.e. land facing) habitats (indicated by stars) on two sampling occasions (i.e. summer and winter). Replicate numbers are shown for each sampling event and site. Open symbols: offshore, closed symbols: nearshore, circles: exposed reef sites, squares: sheltered reef sites; grey: summer, black: winter.
Transect data
Reef substrate was characterized according to live (‘hermatypic corals’ and ‘other live cover’, that is soft corals, macro/micro/turf/calcifying algae, sponges and anemones) and dead (‘bare firm substrate’ and ‘loose substrate’) cover using the line intercept method (Hill & Wilkinson 2004) in 0.5 m distances along four 20‐m segments (separated by 5 m intervals) of a 100‐m transect at each sampling site. At one of the nearshore and one of the offshore sites, the abundance of C. echinata was counted in 2‐m‐wide belts (Hill & Wilkinson 2004) along four 20‐m transects at both sides of the reefs, that is the exposed (fore‐reef) and sheltered (back‐reef) side (Table S1, Supporting information).
Sample processing
Water samples were filtered onto 0.22‐μm Isopore filters (Millipore) for gravitational determination (Mettler Toledo XS205) of total suspended matter (TSM) and for DNA extraction of the associated microbial community. C. echinata tissue was removed from the coral skeleton using pressurized air. DNA from water and coral samples was extracted according to the manufacturer's instructions using the QIAGEN DNeasy Plant Mini Kit. Variable regions 5 and 6 of the 16S rRNA gene were amplified using the 784F and 1061R primer pair (Andersson et al. 2008) containing barcodes and Roche 454 pyrosequencing adaptors for subsequent library construction as detailed in Bayer et al. (2013b) and Hamady et al. (2008). Polymerase chain reaction (PCR) was prepared using the QIAGEN Multiplex PCR kit with 0.2 μm of each primer and 2 ng DNA for water samples or 30 ng DNA for coral samples plus DNA/RNA‐free water (TEKnova) to a final PCR volume of 25 μL. Temperature cycling profile for amplification was as follows: 95 °C for 15 min followed by 27 cycles of 95 °C for 30 s, 55 °C for 40 s and 72 °C for 40 s, followed by one cycle at 72 °C for 10 min. For each sample, amplifications were performed in triplicate and combined. PCR products for all samples were quantified using a microplate reader (SpectraMax Paradigm; Molecular Devices) and the Qubit Broad Range assay (Invitrogen) prior to pooling of all samples in equal quantities. Sequencing was performed on the Roche 454 FLX platform. Inorganic nutrient concentrations (nitrate + nitrite, nitrite, ammonia, phosphate and silicate) in the filtrate of the water samples were determined using standard colorimetric tests and a QuickChem 8000 (Zellweger Analysis, Inc.) AutoAnalyzer.
Data analysis
Sequencing data were analysed using the open‐source software mothur (Schloss et al. 2009). Sequence reads were split according to barcodes and quality trimmed prior to alignment against the silva database (silva ssu Release 102). Chimeric sequences were removed using the uchime program as implemented in mothur (Edgar et al. 2011) followed by preclustering of the data to 1‐bp difference to compact data and reduce OTUs generated by sequencing errors (Huse et al. 2010). Remaining singletons (i.e. sequences that were only present once across all samples) were removed from the data set yielding samples with a maximum of 12 503 sequence reads (median: 2013, mean 2939 reads per sample). To obtain a minimum of three colony replicates over the categories reef location, sheltered/exposed environment and summer/winter, data were subsampled to 500 sequence reads to allow for the inclusion of samples with low numbers of sequence reads. Detailed information on sequence counts, taxonomic classification and 16S reference amplicon sequences for all OTUs across all samples used in this study is available as supplemental data (Table S2, Supporting information). Sequence raw data determined in this study have been deposited in the NCBI Sequence Read Archive under accession no. PRJNA277291.
Bacterial assemblages associated with reef water and coral specimens were tested for differences between shelf sites (‘offshore’ vs. ‘nearshore’), exposures (‘exposed’ vs. ‘sheltered’) and time of year (‘summer’ vs. ‘winter’, referenced as ‘season’ in the following) using permutation multivariate analysis of variance (permanova). Here, all fixed factors (‘site’, ‘exposure’ and ‘season’) were nested according to hierarchy, and 999 permutations of residuals were conducted based on Bray–Curtis distances between samples using the primer‐e software with the permanova+ add‐on package (Clarke & Gorley 2006). Environmental differences in water quality (TSM, nutrients, temperature) between shelf sites (‘offshore’ vs. ‘nearshore’), exposures (‘exposed’ vs. ‘sheltered’) and time of year (‘summer’ vs. ‘winter’) were also identified applying permanova as above, but on Euclidean distances between samples. Here, primer‐e's similarity percentage analysis (simper) on Euclidean distances further revealed the main contributors of the parameters under investigation responsible for site and sampling date differences. Substrate cover data between sites was compared applying the anosim treatment based on Euclidean distances in primer‐e (Clarke & Gorley 2006).
Results
Environmental settings
Water quality (Table 1) between fore‐ and back‐reef environments of near‐ and offshore coral reefs (Fig. 1) differed significantly between summer and winter (P = 0.002) and with distance from shore (P = 0.02), but was similar for exposed and sheltered sides within the same reef locations (i.e. nearshore vs. offshore) (Table 2). Sampling date (i.e. summer vs. winter) differences were mainly driven by temperature, while nearshore and offshore reefs differed in concentration of TSM, and to a lesser extent in temperature (Table 2). Nutrient concentrations did not differ significantly between seasons or sites.
Table 1.
Temperature and concentration of suspended matter and inorganic nutrients (nitrite + nitrate, nitrite, ammonia, phosphate and silicate) at sampling sites of the coral Ctenactis echinata
| Reef name | Shelf | Exposure | Sampling date | Temp (°C) | Total suspended matter (TSM) (mg/L) | Nitrite + nitrate (μm) | Ammonia (μm) | Phosphate (μm) | Silicate (μm) | Nitrite (μm) |
|---|---|---|---|---|---|---|---|---|---|---|
| Inner Fsar | NS | Sheltered | Summer | 33 | 4.27 | 0.50 | 0.65 | 0.08 | 0.56 | 0.07 |
| Inner Fsar | NS | Sheltered | Winter | 25 | 6.20 | 0.38 | 0.34 | 0.10 | 0.55 | 0.05 |
| Inner Fsar | NS | Exposed | Summer | 33 | 3.83 | 0.07 | 0.35 | 0.03 | 0.40 | 0.04 |
| Inner Fsar | NS | Exposed | Winter | 25 | 4.78 | 0.24 | 0.39 | 0.13 | 0.43 | 0.05 |
| Al Quad | NS | Exposed | Summer | 32 | 3.00 | 0.09 | 1.17 | 0.04 | 0.27 | 0.06 |
| Al Quad | NS | Exposed | Winter | 25 | 4.60 | 0.16 | 0.77 | 0.02 | 0.20 | 0.26 |
| Abu Roma | OS | Sheltered | Summer | 30 | 1.20 | 0.19 | 0.18 | 0.09 | 0.35 | 0.09 |
| Abu Roma | OS | Sheltered | Winter | 26 | 1.67 | 0.34 | 0.21 | 0.12 | 0.54 | 0.06 |
| Shib Nazar | OS | Sheltered | Summer | 31 | 3.07 | 0.45 | 0.42 | 0.08 | 0.71 | 0.04 |
| Shib Nazar | OS | Sheltered | Winter | 25 | 4.09 | 0.32 | 0.21 | 0.14 | 0.54 | 0.06 |
| Abu Roma | OS | Exposed | Summer | 30 | 1.11 | 0.84 | 0.15 | 0.10 | 0.49 | 0.06 |
| Abu Roma | OS | Exposed | Winter | 26 | 2.20 | 0.15 | 0.19 | 0.11 | 0.45 | 0.01 |
| Shib Nazar | OS | Exposed | Summer | 31 | 3.07 | 0.18 | 0.19 | 0.07 | 0.68 | 0.04 |
| Shib Nazar | OS | Exposed | Winter | 25 | 4.50 | 0.13 | 0.64 | 0.03 | 0.24 | 0.22 |
NS, nearshore; OS, offshore.
Table 2.
Differences in environmental conditions between habitats and sampling dates of the coral Ctenactis echinata
| permanova | d.f. | SS | MS | Pseudo‐F | Unique permutations | Monte Carlo P‐value |
|---|---|---|---|---|---|---|
| Shelf | 1 | 15.96 | 15.96 | 7.31 | 998 | 0.020 |
| Exposure (shelf) | 2 | 2.29 | 1.14 | 0.52 | 999 | 0.691 |
| Season [exposure(shelf)] | 4 | 144.27 | 36.07 | 16.53 | 999 | 0.002 |
| Residuals | 6 | 13.09 | 2.18 | |||
| Total | 13 | 174 |
| SIMPER | ||||||
|---|---|---|---|---|---|---|
| Summer vs. winter average squared distance = 41.06 | ||||||
| Summer average | Winter average | Av.Sq. Dist. | Sq.Dist/SD | Contrib % | ||
| Temperature (°C) | 31.40 | 25.30 | 37.60 | 2.09 | 91.48 | |
| Nearshore vs. offshore average squared distance = 8.21 | ||||||
|---|---|---|---|---|---|---|
| Nearshore average | Offshore average | Av.Sq. Dist. | Sq.Dist/SD | Contrib % | ||
| TSM (mg/L) | 4.45 | 2.61 | 5.00 | 0.93 | 60.87 | |
| Temperature (°C) | 28.80 | 28.00 | 2.83 | 0.89 | 34.51 | |
Results of the permanova analysis showing differences between ‘shelf’ (i.e. reef locations: nearshore vs. offshore), ‘season’ (i.e. sampling date: summer vs. winter) and ‘exposure’ (i.e. fore‐/back‐reef environment: exposed vs. sheltered). Results of SIMPER analyses showing main factors contributing to a total of >90% of the observed differences between sampling dates and shelf locations, respectively.
Benthic cover composition differed significantly between all locations except between the sheltered sides of nearshore and offshore reefs (all sites P ANOSIM = 0.001, Table S3, Supporting information). Live benthic cover was substantially higher in the exposed offshore reefs compared to all other habitats (Fig. 2). While dead substrate at nearshore and offshore sheltered sites mainly consisted of firm rock, the exposed sides of the nearshore reefs were mainly covered by loose substrate. Importantly, Ctenactis echinata had a distinct distribution pattern and was most abundant on the rocky sheltered sides of the offshore reefs and less present at the exposed sides of the same reefs or at nearshore reef sites (P Kruskal–Wallis = 0.0371, Fig. 2).
Figure 2.
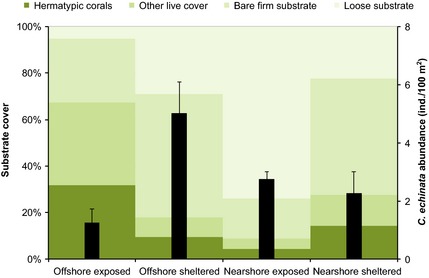
Substrate cover composition (coloured bars) and abundance (black bars) of the coral Ctenactis echinata at different habitats. Offshore sheltered habitats are the species’ preferred habitat and represent distinct environmental conditions. Error bars indicate SE.
Temporal and spatial patterns of microbial communities
To understand microbial assemblage patterns associated with C. echinata over different habitats and sampling dates, we analysed bacterial communities of coral colonies and the surrounding water column via 16S rRNA gene sequencing resulting in a total number of 850 distinct bacterial OTUs at the 0.03 level. Bacterial community profiles of reef water and coral tissue were highly different: only 90 bacterial taxa were encountered in both coral colonies and the water column, while 703 and 237 OTUs were solely associated with coral or water, respectively (Table 3). Further, bacterial communities of water samples showed a highly even distribution independent of sites and conditions (Pielou's evenness J mean = 0.76), whereas evenness of bacterial assemblages associated with corals differed between sites from 0.31 to 0.66 (Pielou's evenness J mean = 0.52) (Table 3). Bacterial assemblages associated with the reef water did not vary between sites, but between summer and winter (Table 4). In contrast, the bacterial diversity of C. echinata (Fig. 3) was highly different between sites as well as between the two sampling dates (i.e. summer vs. winter) (Table 4).
Table 3.
Bacterial profiling of coral and water samples
| Coral/Water | No. of samples | Total no. of OTUs | Average no. of OTUs | Pielou's evenness J | Shannon diversity H′ |
|---|---|---|---|---|---|
| Nearshore | |||||
| Sheltered | |||||
| Summer | 3 | 165 | 55 (±15) | 0.60 (±0.20) | 2.44 (±2.39) |
| Winter | 5 | 304 | 61 (±22) | 0.61 (±0.10) | 2.44 (±2.82) |
| Exposed | |||||
| Summer | 5 | 318 | 64 (±12) | 0.61 (±0.14) | 2.51 (±2.20) |
| Winter | 5 | 312 | 62 (±21) | 0.58 (±0.12) | 2.36 (±2.86) |
| Offshore | |||||
| Sheltered | |||||
| Summer | 8 | 179 | 22 (±4) | 0.31 (±0.19) | 0.99 (±1.71) |
| Winter | 6 | 132 | 22 (±3) | 0.31 (±0.19) | 0.97 (±1.22) |
| Exposed | |||||
| Summer | 3 | 151 | 50 (±15) | 0.66 (±0.11) | 2.58 (±2.40) |
| Winter | 7 | 189 | 27 (±2) | 0.49 (±0.21) | 1.62 (±1.23) |
| Nearshore | |||||
| Sheltered | |||||
| Summer | 1 | 75 | 75 | 0.74 | 3.18 |
| Winter | 1 | 67 | 67 | 0.79 | 3.31 |
| Exposed | |||||
| Summer | 1 | 71 | 71 | 0.78 | 3.31 |
| Winter | 2 | 147 | 74 (±3) | 0.75 (±0.03) | 3.24 (±0.21) |
| Offshore | |||||
| Sheltered | |||||
| Summer | 2 | 120 | 60 (±1) | 0.66 (±0.02) | 2.71 (±0.39) |
| Winter | 2 | 174 | 87 (±8) | 0.79 (±0.08) | 3.51 (±1.36) |
| Exposed | |||||
| Summer | 2 | 147 | 74 (±3) | 0.79 (±0.03) | 3.39 (±0.69) |
| Winter | 1 | 92 | 92 | 0.80 | 3.60 |
| OTUs coral | 703 | ||||
| OTUs water | 237 | ||||
| Shared OTUs | 90 | ||||
| Total no. of OTUs | 850 | ||||
Overview over sample sites and sampling dates, number of samples, number of operational taxonomic units (OTUs) in coral and water, and evenness and diversity indices.
Table 4.
Differences in microbial assemblages associated with reef water and the coral Ctenactis echinata between study sites and sampling dates
| Reef water | d.f. | SS | MS | Pseudo‐F | Unique permutations | Monte Carlo P‐value |
|---|---|---|---|---|---|---|
| Shelf | 1 | 1361 | 1361 | 1.99 | 997 | 0.165 |
| Exposure (shelf) | 2 | 2182 | 1091 | 1.59 | 998 | 0.230 |
| Season [exposure (shelf)] | 4 | 7183 | 1796 | 2.62 | 999 | 0.029 |
| Residuals | 4 | 2737 | 684 | |||
| Total | 11 | 13 782 |
| C. echinata | d.f. | SS | MS | Pseudo‐F | Unique permutations | Monte Carlo P‐value |
|---|---|---|---|---|---|---|
| Shelf | 1 | 18 485 | 18485 | 6.63 | 997 | 0.001 |
| Exposure (shelf) | 2 | 16 703 | 8352 | 2.99 | 998 | 0.001 |
| Season [exposure(shelf)] | 4 | 15 646 | 3912 | 1.40 | 997 | 0.050 |
| Residuals | 34 | 94 836 | 2789 | |||
| Total | 41 | 150 820 |
Results of the permanova analyses showing differences between sampling dates (summer vs. winter) in reef water and coral samples, and between shelf locations (offshore vs. nearshore) and fore‐ and back‐reef environments (i.e. exposed vs. sheltered) in C. echinata samples.
Figure 3.
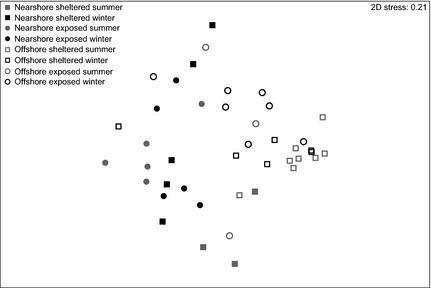
Nonmetric multidimensional scaling plot of bacterial communities associated with Ctenactis echinata samples. Bray–Curtis distances between samples illustrate differences between nearshore and offshore sites as well as between offshore sheltered and offshore exposed environments. Open symbols: offshore, closed symbols: nearshore, circles: exposed reef sites, squares: sheltered reef sites, grey: summer, black: winter.
Coral microbiome composition varies over sites and sampling dates
Of the 703 OTUs associated with coral samples, the 11 most abundant OTUs were encountered on average between 114 and 16 times across all sites and sampling dates. These bacterial taxa accounted for more than 50% of the total bacterial abundance associated with coral samples. Most importantly, the distribution of these abundant taxa differed strongly between sites (Fig. 4). Coral samples from sheltered offshore reef sites were mainly associated with one bacterial taxon (genus Endozoicomonas) representing more than 60% of the total microbial assemblage. The same bacterial taxon was also substantially present in coral samples from the exposed counterparts of the offshore reefs, but was almost entirely missing in samples from nearshore reefs (Fig. 4). The remaining 10 bacterial taxa were present at varying degrees over sites and seasons. For instance, a so far uncharacterized bacterium even at the phylum level (OTU0011) was only present at nearshore exposed sites in summer, but with high read numbers (Table S3, Supporting information). In comparison, other OTUs were more evenly distributed across habitats and sampling dates. All but three OTUs remained unclassified at the genus level. Those identified included another taxon of the genus Endozoicomonas (OTU0019), one taxon of each the genus Vibrio (OTU0003) as well as Photobacterium (OTU0014), both of which belong to the family Vibrionaceae (Table S2, Supporting information).
Figure 4.
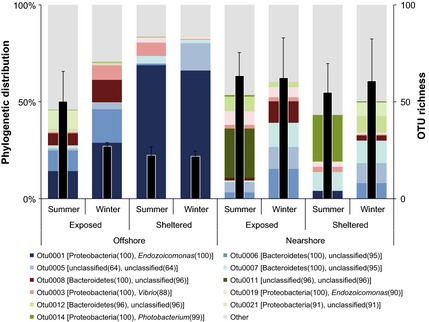
Phylogenetic distribution and operational taxonomic units (OTU) richness of bacteria associated with the coral Ctenactis echinata across different habitats. Stacked bars: phylogenetic affiliation of abundant OTUs. Numbers next to phylum and genus denote bootstrap values of classification. Black bars: average number of OTUs associated with C. echinata. Error bars: SE.
Discussion
The diversity of coral‐associated microbes is controlled by intrinsic (host‐regulated) as well as external (habitat‐regulated) factors (Thompson et al. 2014). Even though a coral host's metabolism contributes to the structure of coral‐associated microbiota (Brown & Bythell 2005), evidence is accumulating that environmental factors such as geographic location, depth, coral and algal cover of the habitat, or elevated temperatures and nutrient concentrations can also influence the coral microbiome (Vega Thurber et al. 2009; Kelly et al. 2014; Pantos et al. 2015). By collecting ecological and molecular data for the coral Ctenactis echinata across different habitats and sampling dates (i.e. summer and winter), we were able to derive the distribution range of this coral species and could compare these data to the host‐associated microbial community.
Our data show that bacterial community composition is indicative of a coral's preferred environment (as derived from coral species abundance) and it changes with distance to it. Where C. echinata is most abundant, the microbiome is highly structured and dominated by a single bacterial taxon in the genus Endozoicomonas. Moving towards habitats where C. echinata is less abundant (i.e. potentially marginal habitats), the microbial community assemblage becomes less structured and more diverse. Most importantly, the bacterial community structure aligns with environmental factors including time of the year, water and substrate quality. As such, we argue that the diversity of C. echinata's microbiome correlates with the environmental preference of that coral species and that its level of organization might reflect the distance to the host's preferred environment.
Our study took place during the course of a year representative of Red Sea conditions (Edwards & Head 1987) without bleaching or disease incidents. Therefore, shifts towards disintegrated microbial assemblages dominated by opportunistic or pelagic taxa, as observed during bleaching or disease (Bourne et al. 2008; Roder et al. 2014), assumingly did not occur and were not visually present. On the molecular level, however, we observed abundance differences of prevailing and rare members of the bacterial community between sampling dates and sites. It is not clear at this point whether these structural differences in the microbial community represent environmental fluctuations or directional adjustments to a more advantageous coral holobiont composition. It is tempting to speculate that changes in the bacterial assembly contribute to phenotypic plasticity by moving the coral holobiont along fitness landscapes (i.e. alternate ‘stable’ states), but further data are needed to unequivocally interrogate such patterns.
More importantly and independent of sampling date influences, patterns of bacterial community structures coincided with the species’ success to prevail at the different sampling locations. C. echinata was mostly encountered in habitats with open rocky substrates and clear water conditions (as found along the sheltered sides of offshore reefs). At these sites, the microbiome of C. echinata is highly structured (i.e. few OTUs make up the majority of bacterial abundance), with one bacterial taxon of the genus Endozoicomonas dominating the bigger part of the bacterial assemblage. With a decrease in rocky substrate availability (as in the exposed sides of the offshore reefs), but similar water quality, this microbial assemblage pattern weakens. Microbial community structure is entirely different in C. echinata situated in environments characterized by an increase in loose substrate and/or turbidity in the water column (due to higher TSM concentrations) as prevalent in nearshore reef habitats. A habitat‐discrete association between host and Endozoicomonas has also been shown for Acropora millepora from the Great Barrier Reef, however, in the opposite direction with nearshore corals holding a stronger structured microbiome and significantly more Endozoicomonas compared to midshelf specimens (Lema et al. 2014). Following the previous line of argument, A. millepora's preferred habitat might therefore resemble nearshore rather than midshore locations.
Considering that the microbial community is vital for a species’ health (Ezenwa et al. 2012), the patterns observed here might hold clues to abundance differences for corals across habitats in the Red Sea and elsewhere. Despite the wide distribution of members of the genus Endozoicomonas associated with marine organisms including corals, gorgonians and sponges, among others (Speck & Donachie 2012; Bayer et al. 2013a,b; Correa et al. 2013; Forget & Kim Juniper 2013; Jessen et al. 2013; La Riviere et al. 2013; Mendoza et al. 2013; Nishijima et al. 2013; Pike et al. 2013; Rodriguez‐Lanetty et al. 2013; Dishaw et al. 2014; Ransome et al. 2014; Morrow et al. 2015), the functional role of this genus is not known. A suggested role is DMSP breakdown (Raina et al. 2009, 2010); however, recent comparative sequencing of Endozoicomonas genomes isolated from three marine invertebrate hosts confirmed the absence of DMSP‐metabolizing genes in this genus (Neave et al. 2014). Other suggested roles include degradation of complex organic carbon sources (La Riviere et al. 2013) or the production of antimicrobial compounds (Bourne et al. 2008), which has been shown for other coral‐associated bacteria (Ritchie 2006). While their precise function is unknown, current data suggest an important role of Endozoicomonas in the coral holobiont.
As with Endozoicomonas, elucidation of the role of other abundant bacteria was not possible as for the majority of OTU sequences functional data are absent. Also, 16S rRNA gene similarity to characterized bacteria was on average low prohibiting further functional inference. At present, meta‐analyses using existing data on microbial abundance data in corals and integrating these with collected environmental parameters to interrogate co‐occurrence are most promising, but still rare. We could not retrieve further information for the 10 most abundant OTUs (besides Endozoicomonas), other than bacteria from the same genera identified in this study were detected in sea water and coral before.
The increasingly diverse microbial assemblages associated with C. echinata sampled outside the species’ preferred habitat indicate a less stable and less structured microbiome, more reminiscent of stressed corals (Bourne et al. 2008; Kellogg et al. 2013; Roder et al. 2014). Interestingly, even for the most abundant OTUs, no taxon was consistently present across all sites and sampling times (Fig. 4). It remains to be determined whether fluctuations in the associated microbiota in different environments are under active host control, environmentally driven or indicative of decreased control of the host over its bacterial symbionts. A recent study by Franzenburg et al. (2013) showed that antimicrobial peptides of Hydra shape species‐specific bacterial associations, but a similar study in corals is lacking.
In conclusion, we show that microbial communities associated with a coral species comprise a variety of bacterial taxa that differ in abundance and diversity across coral host colonies. Microbial abundance differences align to differences in environmental conditions such as time of year, water quality and substrate availability. In habitats where a coral species is successful (i.e. more abundant), its microbial assemblage appears notably more structured and stable compared to less optimal habitats where key bacterial taxa make way for a less structured community, indicating that ecological niche optimization may shape coral microbiome structure. We can further speculate that the availability of an optimal habitat could be significant for the maintenance of a strongly structured microbiome and its loss might be a key to decreases in coral resilience in habitats of degraded quality or in regard to environmental change.
Conflict of interest statement
The authors declare that no conflict of interest exists.
C.R. and C.R.V. designed and conceived the experiments. C.R., M.A. and M.K. generated data. C.R., C.R.V. and T.B. analysed the data. C.R. and C.R.V. wrote the manuscript.
Data accessibility
16S rRNA gene data were uploaded as online Supplementary information. Sequence raw data determined in this study have been deposited in the NCBI Sequence Read Archive under accession no. PRJNA277291.
Supporting information
Table S1. Overview over sampling sites, transect and environmental data and sequence data collection.
Table S2. Sequence counts, taxonomic classification and 16S reference amplicon sequence for all OTUs identified and over all samples.
Table S3. Differences in substrate cover composition between sampling sites. anosim results comparing substrate cover pairwise and over all sampling sites.
Acknowledgements
We would like to thank the KAUST BioScience Core Laboratory for 454 library generation and sequencing. We would also like to thank Christoph Walcher for assistance with coral sampling and transect data generation. Research reported in this publication was supported by the King Abdullah University of Science and Technology (KAUST).
References
- Andersson AF, Lindberg M, Jakobsson H et al (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One, 3, e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T, Arif C, Ferrier‐Pagès C et al (2013a) Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini . Marine Ecology Progress Series, 479, 75–84. [Google Scholar]
- Bayer T, Neave MJ, Alsheikh‐Hussain A et al (2013b) The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue‐associated Endozoicomonas bacteria. Applied and Environmental Microbiology, 79, 4759–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith‐Keune C (2008) Changes in coral‐associated microbial communities during a bleaching event. ISME Journal, 2, 350–363. [DOI] [PubMed] [Google Scholar]
- Brown BE, Bythell JC (2005) Perspectives on mucus secretion in reef corals. Marine Ecology Progress Series, 296, 291–309. [Google Scholar]
- Clarke KR, Gorley RN (2006) PRIMER v6: User Manual/Tutorial. PRIMER‐E, Plymouth. [Google Scholar]
- Correa H, Haltli B, Duque C, Kerr R (2013) Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae . Microbial Ecology, 66, 972–985. [DOI] [PubMed] [Google Scholar]
- Dishaw LJ, Flores‐Torres J, Lax S et al (2014) The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One, 9, e93386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AJ, Head SM (1987) Key Environments: Red Sea. Pergamon Books Ltd, Oxford. [Google Scholar]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB (2012) Animal behavior and the microbiome. Science, 338, 198–199. [DOI] [PubMed] [Google Scholar]
- Forget NL, Kim Juniper S (2013) Free‐living bacterial communities associated with tubeworm (Ridgeia piscesae) aggregations in contrasting diffuse flow hydrothermal vent habitats at the Main Endeavour Field, Juan de Fuca Ridge. MicrobiologyOpen, 2, 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzenburg S, Walter J, Kunzel S et al (2013) Distinct antimicrobial peptide expression determines host species‐specific bacterial associations. Proceedings of the National Academy of Sciences, USA, 110, E3730–E3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias‐Lopez J, Zerkle AL, Bonheyo GT, Fouke BW (2002) Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Applied and Environment Microbiology, 68, 2214–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R (2008) Error‐correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods, 5, 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Wilkinson C (2004) Methods for Ecological Monitoring of Coral Reefs ‐ A Resource for Managers, p. 117. Australian Institute of Marine Science, Townsville, Australia. [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology, 12, 1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen C, Villa Lizcano JF, Bayer T et al (2013) In‐situ effects of eutrophication and overfishing on physiology and bacterial diversity of the red sea coral Acropora hemprichii . PLoS One, 8, e62091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg CA, Piceno YM, Tom LM et al (2013) Comparing bacterial community composition between healthy and White Plague‐like disease states in Orbicella annularis using PhyloChipTM G3 microarrays. PLoS One, 8, e79801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LW, Williams GJ, Barott KL et al (2014) Local genomic adaptation of coral reef‐associated microbiomes to gradients of natural variability and anthropogenic stressors. Proceedings of the National Academy of Sciences, USA, 111, 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman D, Kashman Y, Rosenberg E, Kushmaro A, Loya Y (2006) Antimicrobial activity of Red Sea corals. Marine Biology, 149, 357–363. [Google Scholar]
- Koren O, Rosenberg E (2006) Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Applied and Environment Microbiology, 72, 5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Riviere M, Roumagnac M, Garrabou J, Bally M (2013) Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the Northwestern Mediterranean Sea. PLoS One, 8, e57385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema KA, Willis BL, Bourne DG (2014) Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environmental Microbiology, 16, 3345–3359. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG (2004) Discovery of symbiotic nitrogen‐fixing cyanobacteria in corals. Science, 305, 997–1000. [DOI] [PubMed] [Google Scholar]
- Li J, Chen Q, Zhang S et al (2013) Highly heterogeneous bacterial communities associated with the South China Sea reef corals Porites lutea, Galaxea fascicularis and Acropora millepora . PLoS One, 8, e71301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Q, Long LJ et al (2014) Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Scientific Reports, 4, 7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman RA, Willis BL, Pfeffer C, Bourne DG (2009) Diversities of coral‐associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiology Ecology, 68, 152–163. [DOI] [PubMed] [Google Scholar]
- McFall‐Ngai M, Hadfield MG, Bosch TC et al (2013) Animals in a bacterial world, a new imperative for the life sciences. Proceedings of the National Academy of Sciences, USA, 110, 3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M, Guiza L, Martinez X et al (2013) A novel agent (Endozoicomonas elysicola) responsible for epitheliocystis in cobia Rachycentrum canadum larvae. Diseases of Aquatic Organisms, 106, 31–37. [DOI] [PubMed] [Google Scholar]
- Morrow KM, Bourne DG, Humphrey C et al (2015) Natural volcanic CO2 seeps reveal future trajectories for host‐microbial associations in corals and sponges. ISME Journal, 9, 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatine L, Cernichiari E (1969) Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biological Bulletin, 137, 506–523. [DOI] [PubMed] [Google Scholar]
- Neave MJ, Michell CT, Apprill A, Voolstra CR (2014) Whole‐genome sequences of three symbiotic endozoicomonas strains. Genome Announcements, 2, e00802‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishijima M, Adachi K, Katsuta A, Shizuri Y, Yamasato K (2013) Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. International Journal of Systematic and Evolutionary Microbiology, 63, 709–714. [DOI] [PubMed] [Google Scholar]
- Pantos O, Bongaerts P, Dennis PG, Tyson GW, Hoegh‐Guldberg O (2015) Habitat‐specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix . ISME Journal, doi:10.1038/ismej.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike RE, Haltli B, Kerr RG (2013) Description of Endozoicomonas euniceicola sp. nov. and Endozoicomonas gorgoniicola sp. nov., bacteria isolated from the octocorals Eunicea fusca and Plexaura sp., and an emended description of the genus Endozoicomonas . International Journal of Systematic and Evolutionary Microbiology, 63, 4294–4302. [DOI] [PubMed] [Google Scholar]
- Raina JB, Tapiolas D, Willis BL, Bourne DG (2009) Coral‐associated bacteria and their role in the biogeochemical cycling of sulfur. Applied and Environment Microbiology, 75, 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina JB, Dinsdale EA, Willis BL, Bourne DG (2010) Do the organic sulfur compounds DMSP and DMS drive coral microbial associations? Trends in Microbiology, 18, 101–108. [DOI] [PubMed] [Google Scholar]
- Ransome E, Rowley SJ, Thomas S, Tait K, Munn CB (2014) Disturbance to conserved bacterial communities in the cold‐water gorgonian coral Eunicella verrucosa . FEMS Microbiology Ecology, 90, 404–416. [DOI] [PubMed] [Google Scholar]
- Reshef L, Koren O, Loya Y, Zilber‐Rosenberg I, Rosenberg E (2006) The coral probiotic hypothesis. Environmental Microbiology, 8, 2068–2073. [DOI] [PubMed] [Google Scholar]
- Ritchie KB (2006) Regulation of microbial populations by coral surface mucus and mucus‐associated bacteria. Marine Ecology Progress Series, 322, 1–14. [Google Scholar]
- Roder C, Arif C, Bayer T et al (2014) Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME Journal, 8, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Lanetty M, Granados‐Cifuentes C, Barberan A, Bellantuono AJ, Bastidas C (2013) Ecological Inferences from a deep screening of the Complex Bacterial Consortia associated with the coral, Porites astreoides . Molecular Ecology, 22, 4349–4362. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N (2001) Diversity of bacteria associated with the Caribbean coral Montastrea franksi . Coral Reefs, 20, 85–91. [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N (2002) Diversity and distribution of coral‐associated bacteria. Marine Ecology Progress Series, 243, 1–10. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber‐Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nature Reviews. Microbiology, 5, 355–362. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck MD, Donachie SP (2012) Widespread Oceanospirillaceae bacteria in Porites spp. Journal of Marine Biology, 2012, 7. [Google Scholar]
- Sunagawa S, Woodley CM, Medina M (2010) Threatened corals provide underexplored microbial habitats. PLoS One, 5, e9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JR, Rivera HE, Closek CJ, Medina M (2014) Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Frontiers in Cellular and Infection Microbiology, 4, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R, Willner‐Hall D, Rodriguez‐Mueller B et al (2009) Metagenomic analysis of stressed coral holobionts. Environmental Microbiology, 11, 2148–2163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Overview over sampling sites, transect and environmental data and sequence data collection.
Table S2. Sequence counts, taxonomic classification and 16S reference amplicon sequence for all OTUs identified and over all samples.
Table S3. Differences in substrate cover composition between sampling sites. anosim results comparing substrate cover pairwise and over all sampling sites.
Data Availability Statement
16S rRNA gene data were uploaded as online Supplementary information. Sequence raw data determined in this study have been deposited in the NCBI Sequence Read Archive under accession no. PRJNA277291.


