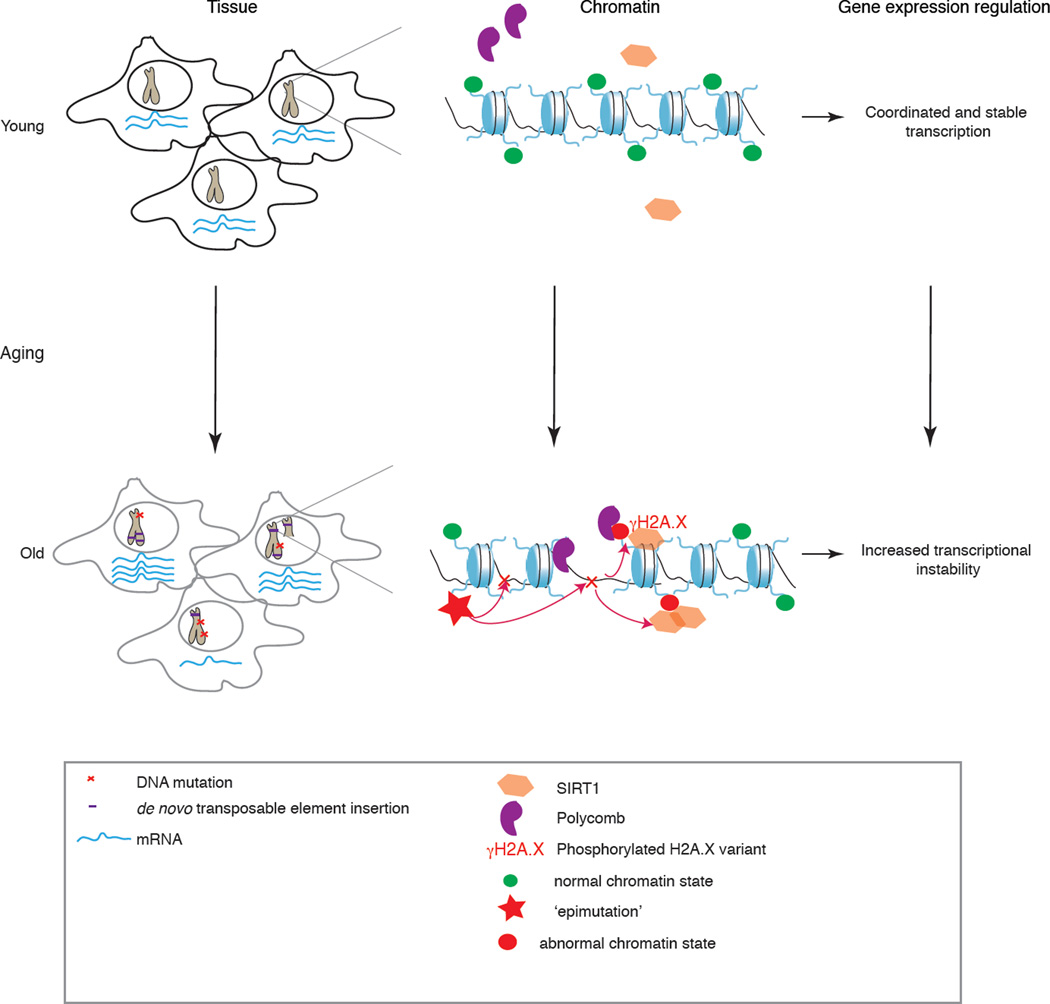
Figure 2. Model for the possible cross-talk between chromatin changes and transcriptional and genomic instability during ageing.
A possible model is the following: in cells from young organisms (top left), transcriptional programs are robustly defined and precise between cells (depicted by consistent mRNA levels). The genomic integrity is maintained (depicted by intact chromosomes), because mutations are rare or correctly repaired. As a result, ‘normal’ chromatin states are found throughout the genome. With increased age (bottom left), transcriptional instability is increased between cells of a tissue (depicted by variable mRNA levels between cells). Genomic instability is also a hallmark of ageing and is increased at both a macro level (for example, aneuploidies depicted by partial chromosome duplication), increased transposable element insertions and, more locally by DNA mutations in the form of single nucleotide mutations, small insertions or deletions (indels). DNA damage can trigger the recruitment of chromatin modifiers, and the acquisition of abnormal chromatin states. Thus, genomic instability could modify the epigenetic landscape of old cells. Reciprocally, aberrant changes in epigenetic marks, known as ‘epimutations’, can further promote the accumulation of DNA mutations in a feedback loop mechanism. The epigenetic changes acquired during ageing could also decrease the transcriptional precision of neighboring genes.