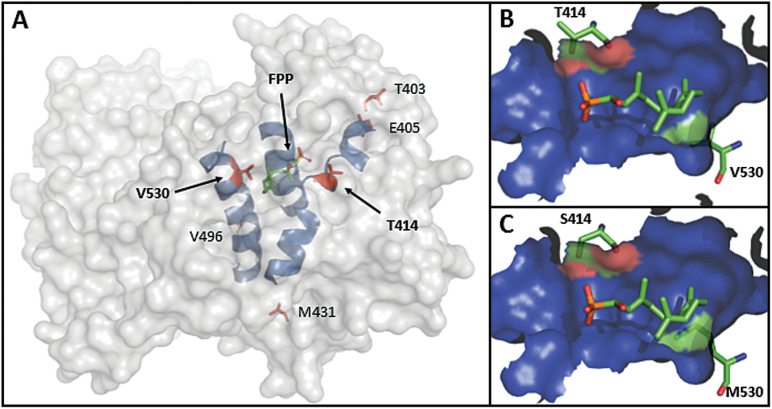
Fig. 2.
A molecular model of the global VvGuaS protein structure based on the TEAS template. (A) A computed enzyme surface is shown as a transparent grey solid, three relevant helices of the secondary structure are visible in blue, and bound FPP, as located in TEAS, is shown centrally located in green (diphosphate group in red). The position of the six amino acid residues that differ between VvPNSeInt and VvGuaS is shown in red and labelled, with T414 and V530 indicated by an arrow. (B) Close-up view of part of the internal active site cavity of VvGuaS and (C) VvPNSeInt. The molecular surface of the internal cavity is shown predominantly in blue, with the FPP substrate and side-chains of amino acid residues 414 and 530 (VvGuaS numbering used) shown as sticks. The contributions of relevant amino acid residues to the formation of the cavity molecular surface are shown using the equivalent colours of the contributing atoms (carbons in green and oxygen in red).