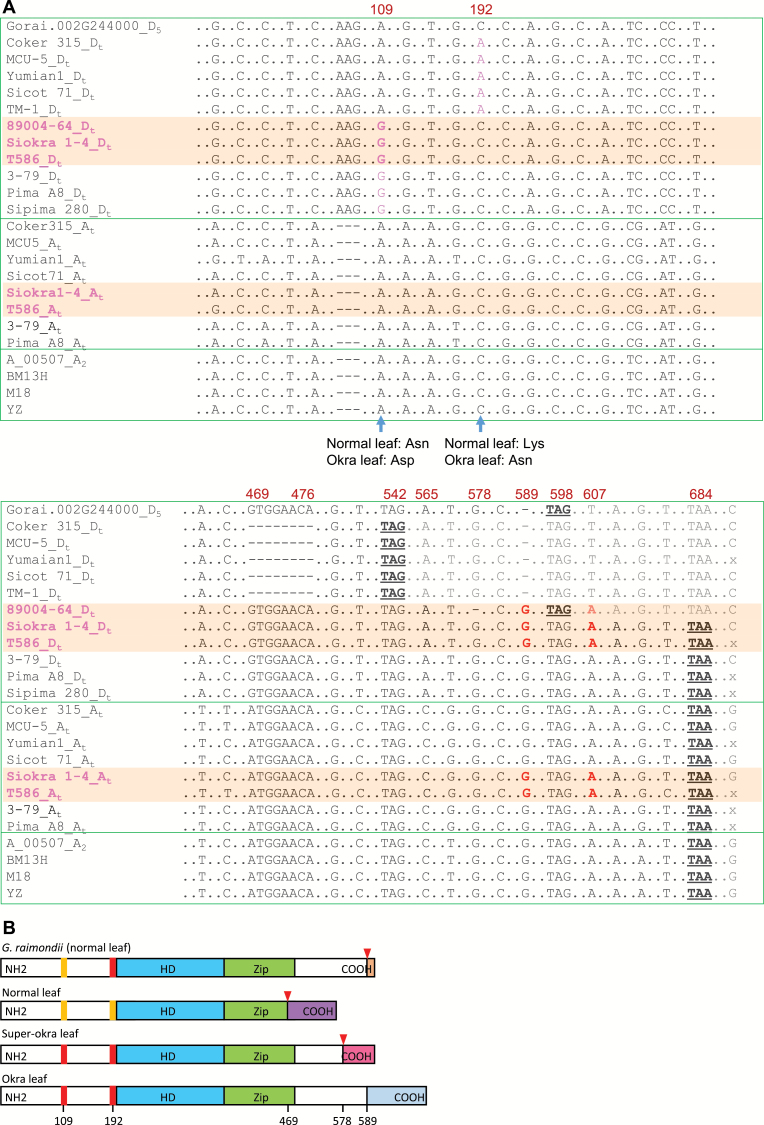
Fig. 3.
Comparison of the nucleotide and protein sequences of the okra leaf gene. (A) Alignment of the coding sequences of Gorai.002G244000 and its orthologues in other cotton species. Only the positions that are polymorphic between any two sequences are shown. Accession name followed by Dt (e.g. Coker 315_Dt) and At represents the Dt and At subgenome allele, respectively. Coker 315, MCU-5, Yumian1, Sicot 71, and TM-1 are normal leaf G. hirsutum accessions; 89004-64, Siokra 1-4, and T586 (shown in pink and highlighted) are superokra leaf or okra leaf G. hirsutum accessions. Pima A8, 3-79, and Sipima 280 are G. barbadense accessions showing the subokra leaf shape. YZ (okra), M18, and BM13H (subokra) are G. arboreum accessions. A dash represent a deletion. Stop codons are shown in bold and underlined. The Dt subgenome SNPs between the normal and okra leaf accessions and the SNPs suggesting the NRHR event are shown in pink and red, respectively. Positions shown on top of the sequences were based on GhOKRA-D t of Siokra 1-4, i.e. Siokra 1-4_Dt. An ‘x’ at the end of some sequences indicates that data were unavailable. (B) Schematic representation of GhOKRA-Dt from G. raimondii and normal, okra, and superokra leaf G. hirsutum accessions. Rectangles represent protein sequences with differences shown in different colours. The numbers at the bottom indicate the nucleotide locations of SNPs or indels. Red triangles indicate the positions of indel(s) that caused a frame shift of the okra leaf allele. HD, homeodomain; Zip, Zip domain.