Figure 4.
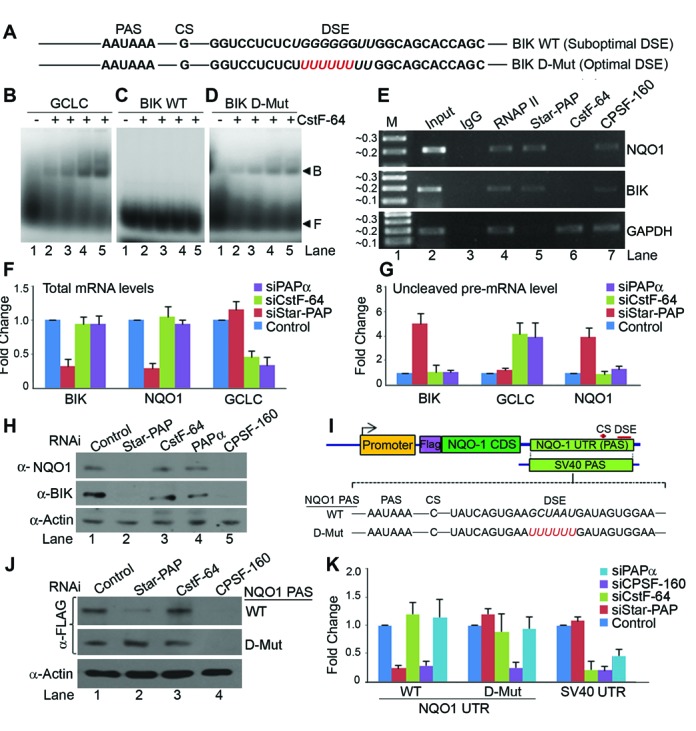
Suboptimal DSE at the 3′-UTR of Star-PAP target mRNAs prevent CstF-64 binding that excludes PAPα from the UTR. (A) BIK UTR sequence indicating the suboptimal DSE region and the insertion of U-rich DSE (UUUUUU) at the UTR. RNA EMSA experiment of CstF-64 with (B) GCLC (C) BIK UTR and (D) mutant BIK with U-rich DSE. F-free probe, B- CstF-64-RNA binary complex. (E) RIP analysis of Star-PAP, CstF-64, CPSF-160 and RNA Pol II on mRNAs as indicated. (F) qRT-PCR analysis of BIK, NQO1 and GCLC mRNA expressions under the conditions as indicated. (G) Uncleaved mRNA measurement by qRT-PCR expressed relative to the total mRNA levels under conditions as in F. (H) Western blot analysis of NQO1, BIK and control α-Actin from lysates of HEK 293 cells after knockdown of Star-PAP, CstF-64, CPSF-160, PAPα or control cells. (I) Reporter constructs as in Figure 3J, showing DSE region and the U-rich DSE insertions. (J) Reporter assay by western blot analysis using anti-FLAG antibody from HEK 293 cells after transfection of the reporter construct under the conditions as indicated. (K) qRT-PCR analysis under the similar conditions as in J.
