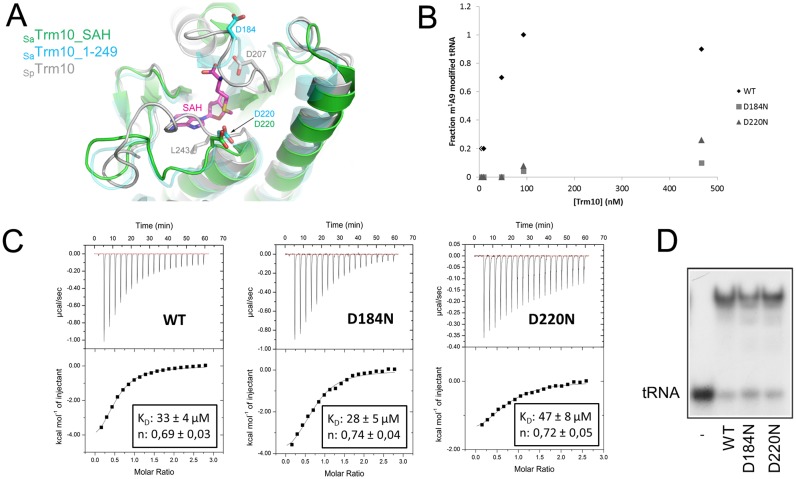
Figure 4.
Role of Asp184 and Asp220 in the catalytic mechanism of SaTrm10. (A) Overlay of the crystal structures of SaTrm10_SAH (green), SaTrm10_1–249 (blue) and SpTrm10 (grey) showing the position of the catalytic residues (Asp184/Asp220 in SaTrm10 and Asp207 in SpTrm10). The bound reaction product SAH is shown as purple sticks. Note that residue Asp184 could not be modelled in the SaTrm10_SAH crystal structure. (B) Methyltransferase activity assay (with in vitro transcribed 32P-labelled SatRNAiMet) comparing the catalytic activity of wild-type SaTrm10 with the one of the D184N and D220N variants at different protein concentrations (and a fixed tRNA concentration). (C) Isothermal titration calorimetry (ITC) assay comparing the binding of SAM to either wild-type SaTrm10 or the D184N or D220N variant. Extra control experiments to determine the heat of dilution and using a protein variant affected directly in the SAM binding pocket can be found in Supplementary Figure S7. (D) Electrophoretic mobility shift assay (EMSA) showing the binding of SatRNAiMet on the wild-type, D184N or D220N SaTrm10 variant.