Abstract
MutY adenine glycosylases prevent DNA mutations by excising adenine from promutagenic 8-oxo-7,8-dihydroguanine (OG):A mismatches. Here, we describe structural features of the MutY active site bound to an azaribose transition state analog which indicate a catalytic role for Tyr126 and approach of the water nucleophile on the same side as the departing adenine base. The idea that Tyr126 participates in catalysis, recently predicted by modeling calculations, is strongly supported by mutagenesis and by seeing close contact between the hydroxyl group of this residue and the azaribose moiety of the transition state analog. NMR analysis of MutY methanolysis products corroborates a mechanism for adenine removal with retention of stereochemistry. Based on these results, we propose a revised mechanism for MutY that involves two nucleophilic displacement steps akin to the mechanisms accepted for ‘retaining’ O-glycosidases. This new-for-MutY yet familiar mechanism may also be operative in related base excision repair glycosylases and provides a critical framework for analysis of human MutY (MUTYH) variants associated with inherited colorectal cancer.
INTRODUCTION
Reactive oxygen species (ROS) present in cells due to aerobic metabolism, inflammatory responses and dysfunctional redox regulation erode genomic integrity by converting normal bases into mutagenic lesions (1–3). The ROS oxidation product 8-oxo-7,8-dihydroguanine (OG) is highly mutagenic due to its ability to mimic thymine (3,4). Indeed, misincorporation of A opposite OG during DNA replication provides the opportunity to introduce G to T transversion mutations in the genome (3,5). MutY adenine glycosylases play a pivotal role in preventing OG-associated mutations by capturing the OG:A mismatch and catalyzing adenine excision as the first step of base excision repair (BER) (3,6). After adenine excision and removal of the abasic site, gap-filling and ligation produce an OG:C base pair that is processed by an OG glycosylase to restore the original G:C through a second round of BER (3). A decade ago, defective repair of OG:A mismatches due to inherited biallelic variations in the human MutY homologue (MUTYH) was linked to colorectal cancer (7), a syndrome now referred to as MUTYH-Associated Polyposis (MAP) (3,8,9). Clinical studies of MAP have progressed at a dramatic pace and over 100 different missense and truncating mutations in MUTYH have been reported to date. However, predicting the magnitude and extent of dysfunction of a particular MUTYH variant and relating this to the severity of the disease remains challenging. Connecting the molecular mechanism and clinical observations would be aided by precise understanding of the chemical steps involved in MutY-mediated repair of OG:A mismatches.
Insight into substrate recognition and adenine excision by MutY has been gleaned from analysis of kinetics of mutated MutY enzymes and alternative substrates, kinetic isotope effect (KIE) studies and structural studies (6,10–16). KIE measurements performed on Escherichia coli (Ec) MutY with a G:A substrate support a DN*AN‡ (SN1) mechanism, featuring a highly reactive oxacarbenium ion intermediate (Figure 1A) (15). In addition, the KIE measurements highlight the importance of protonation of adenine at N7 to enhance its departure, followed by rapid and reversible formation of an oxacarbenium ion intermediate. Finally, irreversible nucleophilic attack at the anomeric carbon leads to formation of an apurinic-apyrimidinic (AP) site product. The crystal structure of a thermophilic homolog, Geobacillus stearothermophilus (Gs) MutY bound to a cleavage resistant substrate analog, arabino 2′-fluoro-2′-deoxyadenosine (FA; this structure is commonly referred to as the Fluorinated Lesion Recognition Complex or FLRC), provides a glimpse after OG:A base pair recognition and adenine base extrusion from the DNA helix into the enzyme active site, prior to N-glycosidic bond hydrolysis (17). This FLRC structure, along with mutagenesis, kinetics and previous structural studies (10,11,13,16), implicated a Glu residue (Glu 43 in Gs MutY) as the acid catalyst and an Asp residue (Asp 144 in Gs MutY) for stabilization of the resulting oxacarbenium ion-like transition state and intermediate (Figure 1A).
Figure 1.
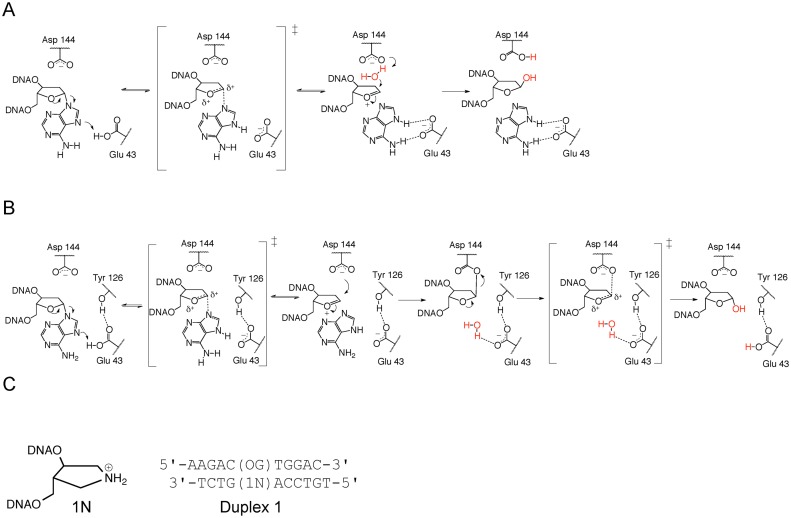
The synthetic analog, 1N, mimics the oxacarbenium ion transition state and intermediate. (A) The SN1 mechanism for MutY proposed on the basis of KIE studies (15) and previous structural studies (16,17) features stabilization of an oxacarbenium ion TS by Asp 144 and nucleophile attack from the 3′ face. (B) The double-displacement mechanism proposed on the basis of this work features two oxacarbenium ion TSs, interception of the first TS by Asp 144 to form a covalent enzyme intermediate and nucleophile attack from the 5′ face facilitated by Glu 43 acting as a general base catalyst. (C) Structure of 1N and its location in Duplex 1 opposite an OG lesion.
A clear molecular picture of how the enzyme generates and stabilizes the predicted chemical intermediates would provide insight into the catalytic strategies used by MutY enzymes. We now report a structure of Gs MutY in complex with a DNA duplex containing an azaribose transition state analog that reveals roles of enzyme residues in facilitating adenine departure and stabilizing the oxacarbenium ion transition state and intermediate. Importantly, the transition state analog structure indicated that the adenine leaving group must exit the active site prior to nucleophilic attack, with net retention of stereochemistry. This structural inference was confirmed using NMR analysis of products produced by MutY in the presence of methanol. Based on these results we propose a two-step mechanism with a covalent MutY-DNA intermediate (Figure 1B). Although different from the currently accepted mechanism (Figure 1A), this two-step mechanism is in accord with all existing structural, biochemical and mutagenesis data. Given the high level of evolutionary conservation among BER glycosylases, it is highly likely that similar catalytic mechanisms are used throughout this important class of enzymes. Indeed, structurally similar active sites for the glycosylases MBD4 and AlkA prompted suggestions that these enzymes may act via a covalent DNA-enzyme intermediate (18,19). The potential impact this revised mechanism holds for cancer therapies targeting DNA repair are discussed.
MATERIALS AND METHODS
Synthesis and purification of DNA oligonucleotides and enzymes
Details are provided in the Supplementary Information. The following sequences were used:
Duplex 1 (5′-TGTCCA X GTCT-3′:3′-CAGGT Y CAGAA-5′), Duplex 2 (5′-AGCTC X TGACT-3′:3′-GCTCGAG Y ACTGACG-5′), Duplex 3 (5′-CTGTAACGGGAGCT X GTGGCTCCATGATCG-3′:3′-GACATTGCCCTCGA Y CACCGAGGTACTAGC-5′) and Duplex 4 (5′-GCGTCCA X GTCTACC-3′:3′-FAM-CGCAGGTYCAGATGG-5′), where X is either A, FA, AP, 1N or THF, Y is either OG or G, and FAM is fluorescein.
Dissociation constant determination and adenine glycosylase assays
Gs MutY glycosylase assays were performed following procedures previously described by our laboratory for Ec MutY (14) and described in the Supplementary Information in more detail. The presence of the methanol adduct was inferred by reduction in the amount of hydrolysis product detected in glycosylase assays. HPLC-based assays allowed for direct detection of methanolysis and hydrolysis products. These reactions contained 10 μM of unlabeled OG:A-containing Duplex 2 with 50–100% excess enzyme (based on total protein) in solutions of 20% methanol, 20 mM Tris pH 7.6, 10 mM EDTA, 0.1 mg/ml BSA and 30 mM NaCl. Reaction aliquots were quenched with sodium hydroxide (0.2 mM) and heated for 5 min at 90°C. The reaction products were separated by HPLC using an anion-exchange column (Dionex DNAPac PA-100 9 × 250 mm) with a linear gradient of ammonium acetate in acetonitrile:water, 9:1 (200 mM to 1.2 M over 25 min). ESI-MS was used to confirm identity of oligonucleotide product peaks observed in the HPLC.
Relative affinity of WT Gs MutY for various analog-containing DNA duplexes was investigated by Electrophoretic Mobility Shift Assay (EMSA) with Duplex 3 as previously described by our laboratory (20,21) and fluorescence polarization experiments with Duplex 4. Details are described in the Supplementary Information.
Crystallization and structure determination
Crystallization, data processing and model refinement are described in the Supplementary Methods. Crystals of wild-type Gs MutY complexed with Duplex 1 DNA containing OG and 1N were obtained by hanging-drop vapor diffusion following micro-seeding (22) and diffracted X-rays to the 2.2-Å resolution limit at the Advanced Light Source SIBYLS beamline 12.3.1 (23). Cross-linking of MutY to DNA was not applied. The final model comprises Gs MutY residues 7–360 (residues 290–294 are missing), 21 nucleotides of Duplex 1 DNA (the 5′ dT is missing in the strand containing 1N), two calcium ions, an iron-sulfur cluster (4 iron (II/III) and 4 sulfur atoms) and 70 water molecules. Coordinates have been deposited in the Protein Data Bank (PDB ID 5DPK).
NMR stereochemical determination
Details of the sample preparation, NMR pulse-sequences and assignments are provided in the Supplementary Information.
RESULTS
Transition State Analog Complex (TSAC) structure determination and overview
Given the considerable precedence for use of cationic nitrogen-containing sugar analogs as tight binding inhibitors for mechanistically similar glycosidases and BER glycosylases (24–32), we reasoned that DNA duplexes containing the positively charged (3R,4R)-4-(hydroxymethyl)pyrrolidin-3-ol (1N) nucleotide opposite OG would be unreactive mimics of the oxacarbenium ion transition states and intermediates, and therefore useful for biochemical and structural studies of MutY (see Figure 1C). Consistent with this hypothesis, dissociation constants (Kd) measured using EMSA reveal extremely high affinity of Gs MutY for OG:1N- and G:1N-containing duplexes (Kd < 5 pM) (Supplementary Table S1). Relative affinity gauged using EMSA (Supplementary Table S1) and fluorescence polarization titrations (Supplementary Figure S1) show that Gs MutY exhibits higher affinity for duplexes containing 1N over those containing the substrate (FA) analog or product (THF) analog. Similar trends in binding affinity for 1N duplexes were also observed with Ec MutY (Supplementary Table S2).
The high affinity of MutY for duplexed DNAs containing 1N prompted crystallization trials of Gs MutY with Duplex 1 DNA containing OG opposite 1N (Figure 1C). This DNA is analogous to the DNA found in the FLRC, the only differences being that 1N replaces FA and a normal dA nucleotide replaces the sulfhydryl-bearing crosslinking nucleotide found in the FLRC (17). Crystals of Gs MutY complexed with Duplex 1 containing 1N diffracted X-rays to the 2.2 Å resolution limit, and indexed to the same space group with comparable unit cell dimensions as previously described for Gs MutY-DNA co-crystals (10,17). Notably, X-ray diffraction quality crystals were obtained using the OG:1N duplex without employing disulfide DNA-enzyme cross-linking as had been used to obtain previous Gs MutY substrate DNA structures. Initial phases were obtained by molecular replacement, and the resulting Transition State Analog Complex (TSAC) structure was refined to yield Rwork / Rfree values of 24.2% / 25.7%. Details for data collection and model refinement statistics are provided in Supplementary Table S3.
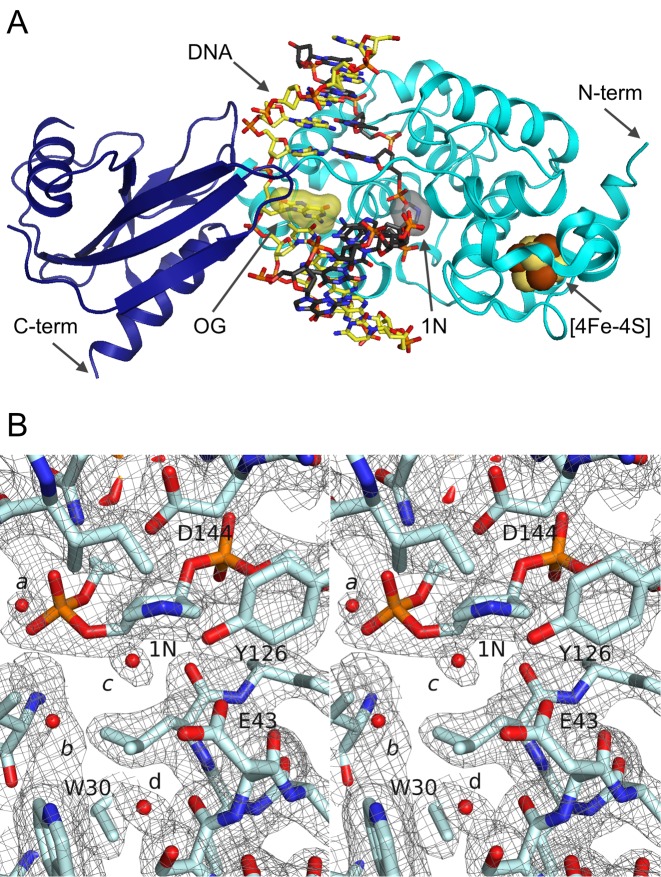
The overall TSAC structure (Figure 2A) is similar to previous structures (10,17). Superposition with the FLRC yields 0.69 Å RMSD for 341 alpha carbons. Key features of the structure include Gs MutY induced remodeling of the DNA resulting in a kink at the lesion site with a DNA bend of 50 degrees as determined by CURVES+ (33,34). The catalytic domain of MutY, harboring the Helix-hairpin-Helix (HhH) DNA binding motif and [4Fe-4S] cluster loop (FCL) motif (35), makes extensive contacts to the 1N-containing strand. The OG-containing strand is contacted by both the C- and N-terminal domains, and the OG base remains within the helical stack with an anti N-glycosidic bond conformation. The 1N nucleotide is rotated out of the helix into the Gs MutY active site pocket. Along with the 1N moiety, the active site harbors at least four ordered water molecules (these active-site waters are labeled a-d in Figure 2B).
Figure 2.
The TSAC structure represents a late transition state. (A) An overview of the TSAC structure shows the OG lesion (yellow surface highlighted) stacked within the distorted DNA helix and its estranged partner, the 1N residue, (black surface highlighted) flipped into the MutY active site. The [4Fe-4S] cluster (brown and yellow spheres) is located in the N-terminal domain. (B) The Gs MutY active site is shown in stereoview with 2|Fo| – |Fc| simulated annealing composite omit electron density (37) contoured at the 1.0 σ level. Coordinated solvent molecules, labelled a-d, occupy the active site. Water c is closest to 1N and likely represents the final nucleophile (see text). Electron density close to water b is consistent with two waters with partial occupancies; however, this region was modeled as a single water so as to avoid over-refinement.
Lack of adenine in the TSAC active site
Previous structures of the N-terminal domain of Ec MutY (11) and Gs MutY bound to DNA containing a reduced AP site (rAP) product analog (10) were obtained with adenine in the active site. In our study, crystals of MutY complexed with 1N containing DNA could be obtained in reactions supplemented with 2.5 mM free adenine; however, electron density maps showed no evidence of adenine in the active site. We think that adenine no longer fits in the active site at the point in the reaction trajectory captured in the TSAC structure. Evidence for this idea comes from superposition of TSAC with the previously reported Gs MutY structure containing OG:rAP with soaked-in adenine (PDB ID 1VRL) (10). These structures are highly comparable (0.46 Å RMSD for 332 alpha carbons). Superposition shows steric clashes between free adenine found in the rAP structure and the 1N ring found in TSAC (Supplementary Figure S2). Furthermore, the active site volume seen in the TSAC is too small to accommodate free adenine. The disposition of atoms and chemical groups in our TSAC structure suggests that the adenine base is not held within the active site after N-glycosidic bond cleavage and formation of the oxacarbenium ion transition state. This is consistent with a report of relatively fast (> 5 min−1) adenine release during the Ec MutY reaction with OG:A duplex substrates (36), which is orders of magnitude faster than release of the AP site DNA product (0.004 ± 0.002 min−1) (14). The structural arrangement observed in the TSAC structure may preclude glycosidic bond re-synthesis; by forcing adenine out of the active site, the enzyme-catalyzed reaction is essentially irreversible. Of note, steric repulsions due to active site reorganization in the MBD4 glycosylase was also suggested to aid in pushing out the cleaved base (19).
Transition state stabilization and nucleophile activation
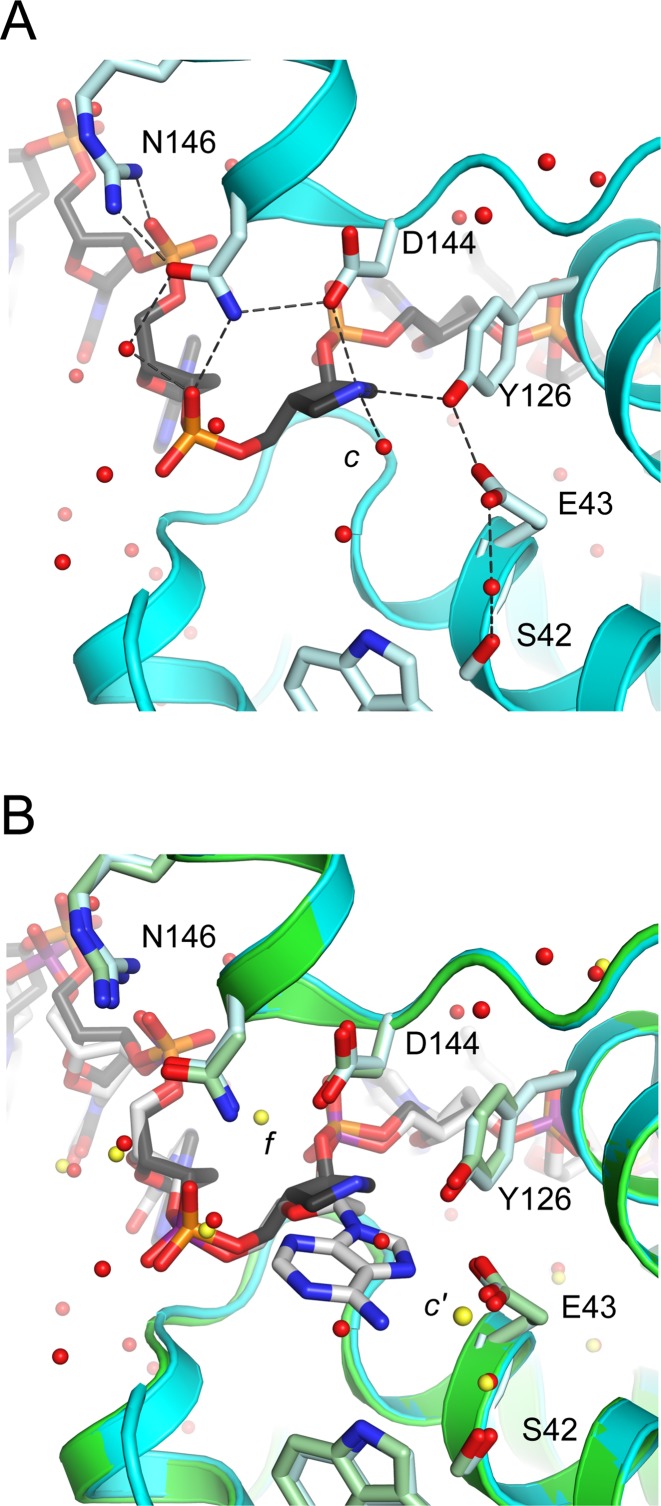
The 1N-containing strand adopts a distorted DNA backbone conformation that positions the transition state analog close to active site residues Asp 144, Glu 43, and Tyr 126 (Figure 3A). Although the current resolution limit warrants caution, we modeled the 1N ring with a 1′-exo conformation because this yielded the best fit to electron density maps compared with 3′-endo or 2′-endo pucker. A similar 1′-exo pucker is seen for 1N in a structure of DNA complexed to uracil DNA glycosylase (PDB 1QRF) (27). The 1N can adopt alternate pucker conformations; a modest 2′-endo pucker is observed for 1N in a structure of DNA complexed with the AlkA DNA glycosylase (PDB 1DIZ) (28). In the TSAC structure, the N1’ atom of 1N participates in hydrogen-bonding and electrostatic interactions from both faces of the azasugar ring. Asp 144 makes a close contact (2.9 Å) to the N1’ atom on the 3′ face. On the 5′ face, robust electron density (5 σ in Fo–Fc discovery maps and 1.5 σ in simulated annealing composite omit maps (37)) indicates the presence of a water molecule (water c in Figure 2B) that is within hydrogen-bonding distance of N1’ (2.8 Å) and the hydroxyl group of Tyr 126 (2.8 Å), and is also in close proximity to the catalytic residue Glu 43 (3.6 Å). In addition to these axial contacts with 1N, an equatorial close contact (3.0 Å) is observed between the N1’ atom of 1N and the hydroxyl group of the highly conserved residue Tyr 126 (Figure 3A). This Tyr 126–1N contact is buttressed by hydrogen-bonding between Tyr 126 and Glu 43.
Figure 3.
Gs MutY makes intimate contacts to 1N in the TSAC structure. Hydrogen bonds are indicated as dotted black lines. (A) Active site residues, Asp (D) 144 and Tyr (Y) 126, in the TSAC structure are within hydrogen-bonding distance to the N1’ atom of 1N. Water molecules (red spheres) include water c which is located 2.8 Å from N1’ of 1N. (B) Overlay of TSAC (cyan) with the substrate analog FLRC structure (green) shows water molecule c is situated on the 5′ face of the sugar ring, the same face from which adenine must depart. Water f (yellow) seen in the FLRC structure, positioned behind Asp 144 and Asn 146, was not observed in the TSAC structure. Water c’ (yellow) seen in the FLRC is closest to the position occupied by water c (red) in the TSAC, but must wait for adenine departure before access to the 5′ face.
The molecular interactions described here for the TSAC, which likely captures the reaction at a late point in the reaction following adenine departure, are similar in many respects to those seen for the FLRC, which captures an early point in the reaction. Both TSAC and FLRC structures show close contact with the sugar ring and Asp 144. Interaction energy with Asp 144 should be maximal with the positively charged oxacarbenium ion intermediate, which is mimicked by the positive charge of the N1’ atom in the TSAC. Both TSAC and FLRC structures show Asp 144 hydrogen-bonded with Asn 146 which is, in turn, hydrogen-bonded with the 5′-phosphate of the 1N (or FA) nucleotide. New interactions seen in the TSAC but not in the FLRC involve Tyr 126, which is within hydrogen-bonding distance (3.0 Å) of the N1’ atom of 1N. In the FLRC, the hydroxyl group of Tyr 126 participates in a hydrogen-bonding network that includes N7 of the substrate adenine and Glu 43. Although not previously considered to be a catalytic residue, Tyr 126 was recently implicated by theoretical modeling of the MutY-catalyzed reaction (38). To test the importance of Tyr 126, we compared single-turnover kinetics for wild-type and the single amino-acid substitution variant with Phe replacing Tyr at position 126 and found a 260-fold slower rate of adenine removal (Supplementary Figure S3), corroborating the importance of structural features involving Tyr 126 seen here in the TSAC.
Another key difference from what is observed in the FLRC structure involves the position of solvent molecules within the active site. We propose that water c in the TSAC structure is positioned favorably for nucleophilic attack from the 5′ face after oxacarbenium ion formation (Figure 3A). The TSAC structure is devoid of water molecules in the vicinity of Asp 144 and Asn 146. A water seen in this location in the FLRC (water f in Figure 3B) had been proposed to attack from the 3′ face (17). Catalytic base groups are located on both 3′ and 5′ faces. Asp 144 could accept a proton during nucleophile attack from the 3′ face, as has been proposed (17); Glu 43 is within 3.4 Å of water c and is therefore the most likely catalytic base to facilitate attack from the 5′ face. If water c is the nucleophile, this would predict retention of configuration at C1’. We realize that this trajectory for nucleophile approach requires that adenine exits the active site before water gains access to the 5′ face, entailing a relatively long period of time (μs) for diffusion. The oxacarbenium ion is not expected to persist on this time scale; therefore, based on precedence set by retaining O-glycosidases (39), we suggest that the oxacarbenium ion intermediate is transiently converted to a more stable covalent enzyme-DNA intermediate during adenine departure.
Detecting transglycosylation activity
To distinguish between nucleophilic attack from the 5′ face verses 3′ face, we sought to determine the product stereochemistry of the Gs MutY-catalyzed reaction. Anomerization of the AP site product at C1’ results in loss of stereochemical information of the MutY-catalyzed reaction. The stereochemistry may be determined, however, by capture of the oxacarbenium ion intermediate with methanol to form a stable cyclic acetal product. For instance, the 2′-deoxynucleotide hydrolase, RCL, has been shown to exhibit glycosyl transfer activity to methanol (40). In contrast, AMP nucleosidase activates a water nucleophile that apparently cannot be replaced by methanol (41). These examples illustrate that the ability to accept an alternative nucleophile depends highly on features of the transition state for a given glycosyl hydrolase (42,43). Though use of methanol as the nucleophile during base-excision repair has not previously been reported, two alkyladenine DNA glycosylases have been shown to catalyze O-glycoside bond synthesis using an AP site-containing duplex and solvent alcohols (44).
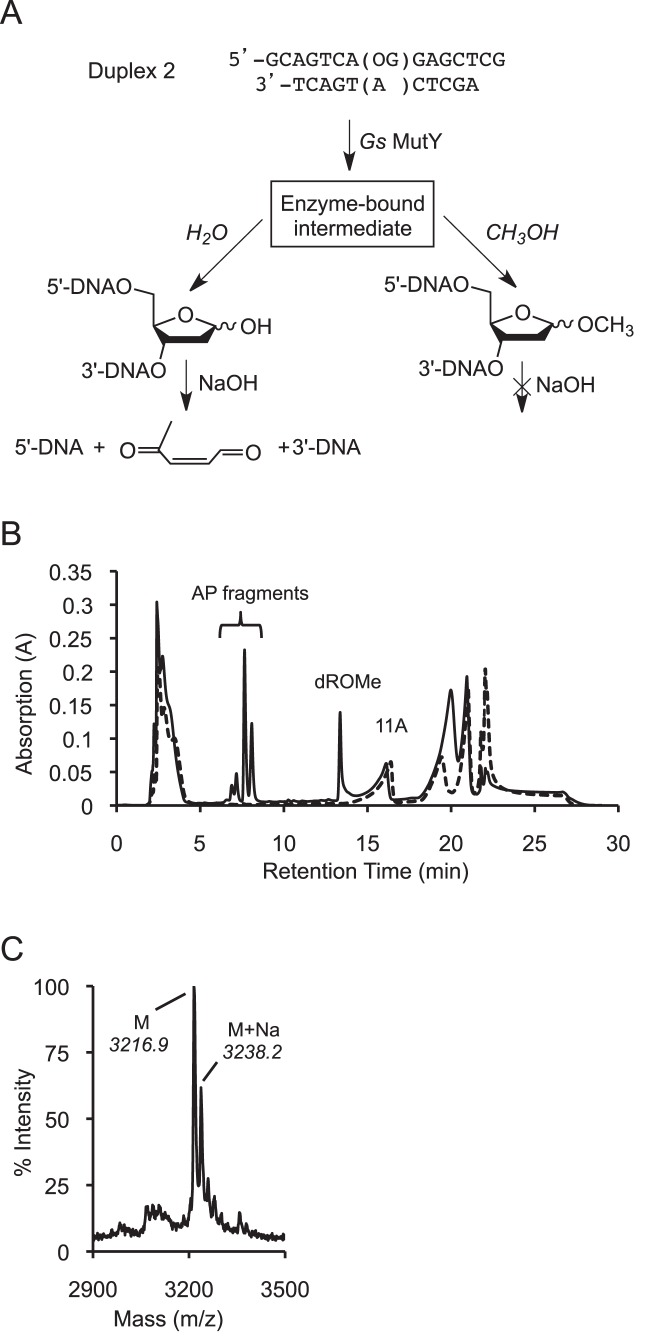
Initial assays showed that the rate of AP site accumulation catalyzed by Gs MutY was similar in the presence or absence of 20% methanol, and that methanol-containing reactions yielded less AP site product, indirectly suggesting formation of a methanolysis product (Supplementary Figure S4). These initial experiments detected the product of hydrolysis but not the product of methanolysis. To directly monitor both hydrolysis and methanolysis products generated by Gs MutY, we used an OG:A DNA substrate (Duplex 2) that positions the OG lesion and target A within 15- and 11-nucleotide sequences, respectively (Figure 4A). In reactions with excess Gs MutY containing methanol and quenched with sodium hydroxide (14), β and δ-elimination of the AP site product produced smaller DNA fragments while the O-methyl-2′-deoxyribonucleotide (dROMe)-containing strand was resistant to sodium hydroxide and therefore remained intact. The various oligonucleotides were resolved using high-performance liquid chromatography (HPLC), and the unique peak for the dROMe-containing oligonucleotide was confirmed by MALDI-MS (Figure 4B and C). Intensities of the HPLC peaks indicate that both the methanolysis (dROMe) and hydrolysis (AP) products were formed in parallel with similar rates (Supplementary Figure S5). In addition, Gs MutY was not able to catalyze formation of the dROMe product starting with an AP site-containing product duplex (Supplementary Figure S6). The ratio of dROMe:AP products formed (25:75) was similar to the solvent composition ratio (20:80 methanol:water) in the reaction mixture, further indication that water and methanol are nearly interchangeable. Importantly, the ability to accept methanol as a nucleophile and the lack of strong solvent-preference suggests that the transition state for the MutY-catalyzed adenine hydrolysis reaction does not involve participation of the water (or methanol) nucleophile. Taken together these results support formation of the dROMe site from an enzyme-bound intermediate generated during MutY-catalyzed adenine excision that proceeds with a highly similar mechanism to that of the native hydrolysis reaction.
Figure 4.
Formation of methyl O-glycoside site in DNA. (A) The hydrolysis product AP site (left path) undergoes β/δ-elimination upon treatment with sodium hydroxide (NaOH). Conversely, the methanolysis product (right path) is resistant to cleavage by NaOH. (B) HPLC traces for methanolysis reactions with Gs MutY (solid) and without enzyme (dashed) indicate baseline separation of all substrates and products and verify that all products are generated by an enzyme-catalyzed reaction. (C) MALDI-MS confirms the presence of a dROMe-oligomer (predicted 3213.1 Da, measured 3216.9 Da, +3.8 offset).
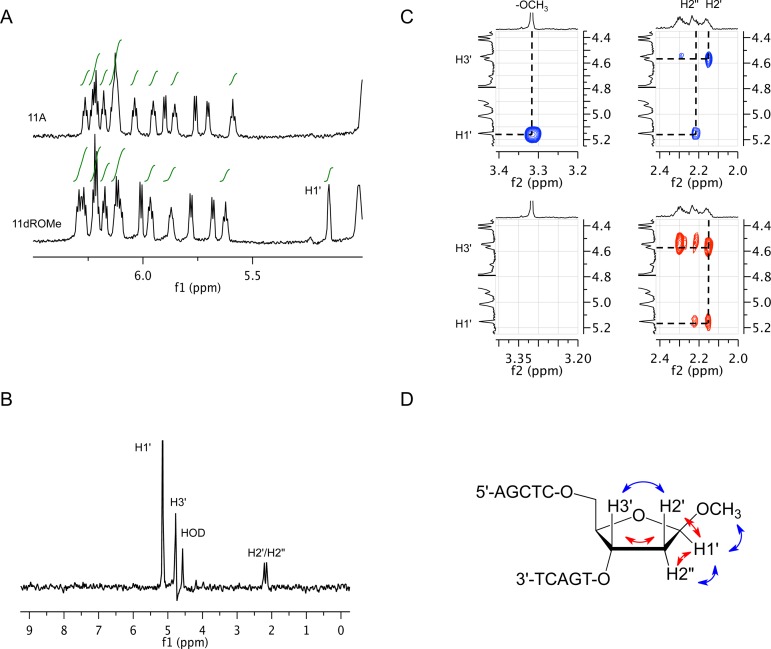
Determination of stereochemistry
The stereochemistry of the dROMe 11-nt product was determined by 1H nuclear magnetic resonance (NMR) spectroscopy. A unique up-field shifted resonance at 5.15 ppm in the anomeric region of the product spectrum was identified as the H1’ of the dROMe O-glycoside (Figure 5A). TOCSY experiments identified the associated H2’/H2’' and H3’ resonances of the dROMe nucleotide (Figure 5B). NOESY and COSY spectra were used to confirm the resonance assignments (Supplementary Tables S4 and S5). Key cross-peaks observed in the NOESY (blue) and COSY (red) spectra provided the basis for the assignment of dROMe stereochemistry at C1’ (Figure 5C and D). The proton resonance at 2.15 ppm was assigned as the H2’ based on a strong NOE with the resonance at 4.57 ppm (H3’). This NOE also necessitated assignment of the resonance at 2.21 ppm as the H2’' that in turn exhibited strong NOEs with resonances for H1’ (5.15 ppm) and -OCH3 (3.32 ppm). In the identical COSY spectral region, the crosspeaks observed provide corroboration of these assignments. The NMR results are consistent with product DNA containing a single anomer of O-methyl 2′-deoxyriboside in the beta configuration, with H1’ on the same ring face as H2’'. The opposite anomeric configuration, alpha, would have provided strong NOE crosspeaks between H1’ and H2’, which were not observed. The chemical shift and J coupling constants of the dROMe methyl O-glycoside site in DNA were also consistent with NMR analysis of the corresponding beta methyl 2′-deoxyribonucleoside. These results strongly support an enzyme-catalyzed methanolysis reaction that occurs with net retention of stereochemistry.
Figure 5.
NMR assignment of beta-configuration for the dROMe nucleotide in the methanolysis product DNA. (A) One-dimensional 1H NMR spectra of the substrate and product oligonucleotides are shown. The H1’ of dROMe is indicated at 5.15 ppm in the product spectrum. (B) The panel shows dROMe sugar proton assignments from a representative TOCSY. (C) 2D NOESY (blue) and COSY (red) show spatial relationship and connectivity. (D) The panel diagrams observed NOESY (blue) and COSY (red) correlations on the structure of the dROMe nucleotide that establish the beta configuration.
DISCUSSION
Structure determination, mutagenesis, kinetic analysis and the discovery that MutY is a retaining glycosylase prompt necessary revisions of the mechanisms previously proposed for MutY based on the FLRC crystal structure and KIE studies (Figure 1A) (15,17). Collectively, the TSAC crystal structure and insights from methanolysis reactions indicate that Gs MutY removes adenine from an OG:A-containing duplex with net retention of stereochemical configuration at the anomeric center. The β-methyl O-glycoside product is proposed to form via a double displacement mechanism at C1’, featuring a covalent enzyme-glycosyl intermediate resulting from attack of the oxacarbenium ion intermediate by an amino acid residue, most likely Asp 144 (Figure 1B). Recent theoretical modeling of the Gs MutY–catalyzed reaction suggested a partial nucleophilic attack by Asp 144 during formation of the oxacarbenium ion intermediate (38). In these calculations, the adenine base was restrained within the active site after N-glycosidic bond cleavage resulting in prediction of formation of the alpha-AP site product, rather than the observed beta anomer. Classical mechanisms of retaining O-glycosidase hydrolases involve two inverting steps and formation of a glycosyl-enzyme intermediate (39,45–47). With respect to similar catalytic mechanisms, the constellation of the catalytic Asp and Glu residues, and in some cases a Tyr residue, many glycoside hydrolases resemble MutY (39). Covalent intermediates have also been proposed for two nucleoside hydrolases based on retention of β-stereochemistry in methanolysis reactions (40,48). With the considerable precedence established by O-glycosidases, it is reasonable to consider ‘classic’ double-displacement for BER glycosylases.
Updated mechanism for MutY
Based on our results and mechanistically similar enzymes that retain stereochemistry, we propose an updated mechanism for MutY (Figure 1B). After substrate recognition and rotation of adenine into the active site pocket, adenine is protonated at N7 by Glu 43 and the N-glycosyl bond is broken via DN*AN‡ (SN1) mechanism as indicated by the KIE measurement (15). The resulting oxacarbenium ion intermediate is attacked by Asp 144 and forms a covalent enzyme intermediate that provides time for adenine departure and nucleophile arrival. During the subsequent hydrolysis step, the water nucleophile approaches C1’ from the 5′ face of the sugar ring and displaces Asp 144, proceeding through a second oxacarbenium ion-like transition state. Tyr 126 participates in a sustained hydrogen bond network with Glu 43 and electrostatically stabilizes the oxacarbenium ion-like transition states for both the base excision and hydrolysis steps. Activation of the water nucleophile in the second TS is likely unnecessary due to the ability of both methanol and water to serve as nucleophiles. In addition, the high reactivity of the oxacarbenium ion intermediate in an SN1 mechanism removes reliance on nucleophile activation in the TS. Based on the positioning of Glu 43 in the TSAC structure we suggest that Glu 43 serves as the proton acceptor and general base catalyst. The double-displacement with retention mechanism described here for MutY is in accord with all existing structural, biochemical and mutagenesis data.
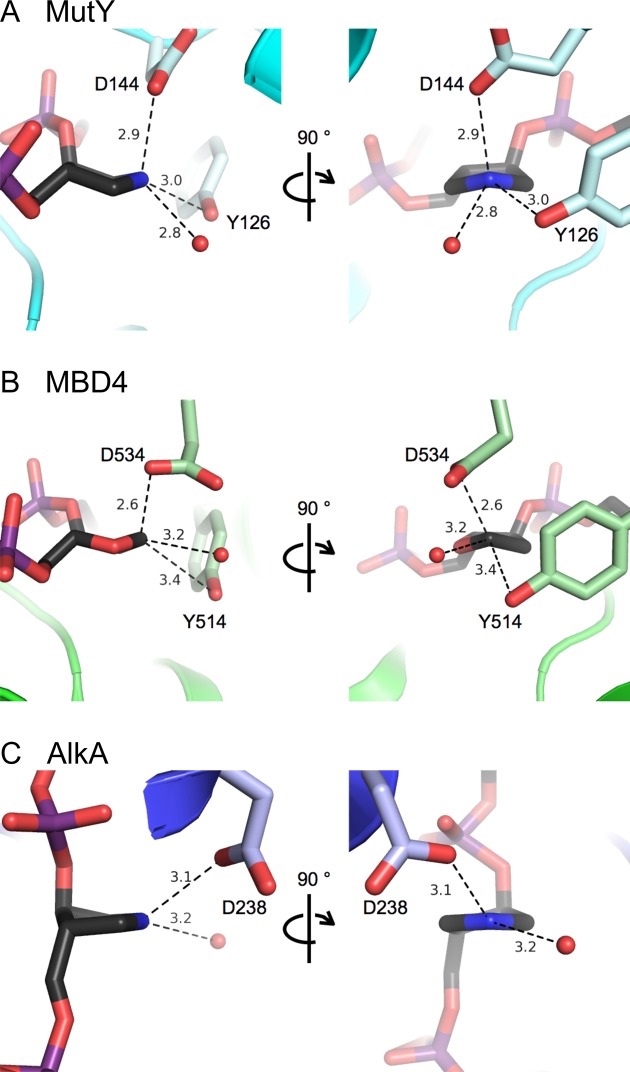
MutY is a member of the BER superfamily of DNA glycosylases, suggesting that other superfamily members use a similar mechanism. Figure 6 compares active sites for MutY and two other BER DNA glycosylases. Notably, structural studies of an intermediate formed during crystallization of MBD4 glycosylase and a G:T-containing duplex were suggestive of a covalent complex between the enzyme and an unhydrolyzed AP site (Figure 6B) (19). In addition, the structure of AlkA bound to a 1N-containing duplex indicated a direct hydrogen bond contact between an Asp residue and N1’, leaving no room for a catalytic water molecule to approach from the 3′ face of the ring (Figure 6C) (28). Indeed, in both reports the authors suggest that a potential alternative mechanism for these glycosylases may be one similar to that utilized by the O-glycosidases (19,28). Moreover, BER glycosylases may exhibit both ‘retaining’ and ‘inverting’ mechanisms, like their O-glycosidase cousins. Subtle differences in the mechanisms used may be related to the nature of the damaged base(s) targeted by a given glycosylase and the ease with which these damaged bases are removed. Doubtless, this double-displacement with retention of stereochemistry mechanism for MutY will prompt similar and additional studies of BER glycosylase mechanisms.
Figure 6.
Comparison of BER DNA glycosylase active sites. Active sites are shown for MutY in TSAC (this work), MBD4 bound to an unhydrolyzed abasic site (19) and AlkA with a 1N TS analog (28). In all three structures, an Asp (D) residue makes close and apparently stabilizing interactions with the TS mimic or abasic site and potential nucleophiles approach from the 5′ face or equatorially.
Implications in human disease
This work also illuminates the structural and functional roles of Gs MutY active site residues (in particular, Glu 43, Tyr 126, Asp 144 and Asn 146) in stabilizing an oxacarbenium ion transition state and participating in covalent catalysis. Knowledge of the catalytic roles of residues in MutY is useful in predicting the potential functional consequences of MAP variants. For example, N224S MUTYH is a rare variant associated with colorectal cancer (49) that corresponds to mutation of the active site residue Asn 146 in Gs MutY (Figure 3). Asn 146 is involved in hydrogen-bonding to both Asp 144 and the phosphate backbone, and therefore mutation to Ser would be anticipated to disrupt this intricate hydrogen-bond network and result in reduced enzyme activity. A plethora of MUTYH missense and truncating variants have been identified (>100) and this number is likely to increase with the rapid expansion of genome sequencing. Many missense variants designated as ‘variants of uncertain significance’ present challenges to clinicians in diagnosis of MAP and further underscores the need for detailed understanding of MUTYH structure-function correlations. In future work, structural studies of cancer-related variants bound to both the substrate and TS analogs would provide insight into how specific mutations alter multiple steps in damage recognition and catalysis to explain underlying causes of MAP.
The mechanistic insight provided herein also provides a basis for the design of a potentially novel class of pharmaceuticals targeted at BER glycosylases. Knowledge of the reaction mechanism and transition states in enzymes that are mechanistically similar to DNA glycosylases has allowed for the development of highly specific and potent small molecule inhibitors to important medicinal targets (50). Notably, a series of purine nucleoside phosphorylase inhibitors, called immucillins, were introduced into preclinical and clinical trials that led toward the development of a new cancer therapeutic (51). Since many cancer cells have compromised DNA repair, targeting the intact DNA repair pathways has shown promise in ‘pushing cancer over the edge’ (52,53). Recently, a new class of inhibitors that target the d(OG)TP hydrolase MTH1 selectively eradicate cancer cells by exploiting their inherent property of high oxidative stress (54,55). These examples suggest that inhibitors developed to target DNA repair glycosylases, such as MUTYH and hOGG1, may be useful anticancer drugs (52).
ACCESSION NUMBERS
Coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5DPK).
Supplementary Material
Acknowledgments
We thank the beamline staff, especially Dr Scott Classen and Dr Kenneth Frankel, for assistance. We thank Dr Greg Verdine for providing a Gs MutY expression construct and Dr Vern Schramm and Dr Setu Roday for providing a 1N-containing oligonucleotide that was used for initial binding tests with Ec MutY. We also thank Dr Jerry Dallas and Dr Jim Ames for their help and advice with the NMR experiments. Lastly we thank Dr Marvin Makinen (University of Chicago) for suggesting the idea of covalent catalysis at an early stage of this project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [CA067985 to S.S.D., GM067994 to M.P.H., GM08537 and CA093247 to V.L.O.]; in part by the U.S. Department of Energy (DOE) program Integrated Diffraction Analysis Technologies (IDAT) (to SIBYLS beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory in part); DOE program Molecular Assemblies Genes and Genomics Integrated Efficiently (MAGGIE) [Contract Number DE-AC02–05CH11231]. The NMR facility at UC Davis is supported in part by NSF grant DBIO 722538. Funding for open access charge: National Institutes of Health (NIH) [CA069875].
Conflict of interest statement. None declared.
REFERENCES
- 1.Cadet J., Douki T., Ravanat J.-L. Oxidatively generated base damage to cellular DNA. Free Radic. Biol. Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Delaney S., Jarem D.A., Volle C.B., Yennie C.J. Chemical and biological consequences of oxidatively damage guanine in DNA. Free Radic. Res. 2012;46:420–441. doi: 10.3109/10715762.2011.653968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grollman A.P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 5.van Loon B., Markkanen E., Hubscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxoguanine. DNA Rep. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 6.David S.S., Williams S.D. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., et al. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 8.Wallace S.S., Murphy D.L., Sweasy J.B. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazzei F., Viel A., Bignami M. Role of MUTYH in human cancer. Mutat. Res. 2013;743–744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Fromme J.C., Banerjee A., Huang S.J., Verdine G.L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y., Manuel R.C., Arvai A.S., Parikh S.S., Mol C.D., Miller J.H., Lloyd S., Tainer J.A. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 12.Michelson A.Z., Rozenberg A., Tian Y., Sun X., Davis J., Francis A.W., O'Shea V.L., Halasyam M., Manlove A.H., David S.S., et al. Gas-phase studies of substrates for the DNA mismatch repair enzyme MutY. J. Am. Chem. Soc. 2012;134:19839–19850. doi: 10.1021/ja309082k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinkmeyer M.K., Pope M.A., David S.S. Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays. Chem. Biol. 2012;19:276–286. doi: 10.1016/j.chembiol.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porello S.L., Leyes A.E., David S.S. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 15.McCann J.A., Berti P.J. Transition-state analysis of the DNA repair enzyme MutY. J. Am. Chem. Soc. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 16.Parikh S.S., Mol C.D., Hosfield D.J., Tainer J.A. Envisioning the molecular choreography of DNA base excision repair. Curr. Opin. Struct. Biol. 1999;9:37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 17.Lee S., Verdine G.L. Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18497–18502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollis T., Ichikawa Y., Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA Glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto H., Zhang X., Cheng X. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012;40:8276–8284. doi: 10.1093/nar/gks628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porello S.L., Williams S.D., Kuhn H., Michaels M.L., David S.S. Specific recognition of substrate analogs by the DNA mismatch repair enzyme MutY. J. Am. Chem. Soc. 1996;118:10684–10692. [Google Scholar]
- 21.Chepanoske C.L., Porello S.L., Fujiwara T., Sugiyama H., David S.S. Substrate recognition by Escherichia coli MutY using substrate analogs. Nucleic Acids Res. 1999;27:3197–3204. doi: 10.1093/nar/27.15.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergfors T. Seeds to crystals. J. Struct. Biol. 2003;142:66–76. doi: 10.1016/s1047-8477(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 23.Classen S., Hura G.L., Holton J.M., Rambo R.P., Rodic I., McGuire P.J., Dyer K., Hammel M., Meigs G., Frankel K.A., et al. Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 2013;46:1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schramm V.L. Enzymatic transition states, transition-state analogs, dynamics, thermodynamics, and lifetimes. Annu. Rev. Biochem. 2011;80:703–732. doi: 10.1146/annurev-biochem-061809-100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horne G., Wilson F.X., Tinsley J., Williams D.H., Storer R. Iminosugars past, present and future: medicines for tomorrow. Drug Discov. Today. 2011;16:107–118. doi: 10.1016/j.drudis.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 26.Chu A.M., Fettinger J.C., David S.S. Profiling base excision repair glycosylases with synthesized transition state analogs. Bioorg. Med. Chem. Lett. 2011;21:4969–4972. doi: 10.1016/j.bmcl.2011.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianchet M.A., Seiple L.A., Jiang Y.L., Ichikawa Y., Amzel L.M., Stivers J.T. Electrostatic guidance of glycosyl cation migration along the reaction coordinate of uracil DNA glycosylase. Biochemistry. 2003;42:12455–12460. doi: 10.1021/bi035372+. [DOI] [PubMed] [Google Scholar]
- 28.Hollis T., Ichikawa Y., Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y.L., Ichikawa Y., Stivers J.T. Inhibition of uracil DNA glycosylase by an oxacarbenium ion mimic. Biochemistry. 2002;41:7116–7124. doi: 10.1021/bi025694y. [DOI] [PubMed] [Google Scholar]
- 30.Werner R.M., Stivers J.T. Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: evidence for an oxocarbenium ion-uracil anion intermediate. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 31.Berti P.J., McCann J.A. Toward a detailed understanding of base excision repair enzymes: transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem. Rev. 2006;106:506–555. doi: 10.1021/cr040461t. [DOI] [PubMed] [Google Scholar]
- 32.Gloster T.M., Davies G.J. Glycosidase inhibition: assessing mimicry of the transition state. Org. Biomolec. Chem. 2010;8:305–320. doi: 10.1039/b915870g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchet C., Pasi M., Zakrzewska K., Lavery R. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 2011;39:W68–W73. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thayer M.M., Ahern H., Xing D., Cunningham R.P., Tainer J.A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCann J.A., Berti P.J. Adenine release is fast in MutY-catalyzed hydrolysis of G:A and 8-Oxo-G:A DNA mismatches. J. Biol. Chem. 2003;278:29587–29592. doi: 10.1074/jbc.M212474200. [DOI] [PubMed] [Google Scholar]
- 37.Hodel A., Kim S.-H., Brünger A.T. Model bias in macromolecular crystal structures. Acta Crystallogr. 1992;A48:851–858. [Google Scholar]
- 38.Kellie J.L., Wilson K.A., Wetmore S.D. Standard role for a conserved aspartate or more direct involvement in deglycosylation? An ONIOM and MD investigation of adenine-DNA glycosylase. Biochemistry. 2013;52:8753–8765. doi: 10.1021/bi401310w. [DOI] [PubMed] [Google Scholar]
- 39.Zechel D.L., Withers S.G. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000;33:11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- 40.Doddapaneni K., Zahurancik W., Haushalter A., Yuan C., Jackman J., Wu Z. RCL hydrolyzes 2′-deoxyribonucleoside 5′-monophosphate via formation of a reaction intermediate. Biochemistry. 2011;50:4712–4719. doi: 10.1021/bi101742z. [DOI] [PubMed] [Google Scholar]
- 41.Parkin D.W., Schramm V.L. Catalytic and allosteric mechanism of AMP nucleosidase from primary, beta-secondary, and multiple heavy atom kinetic isotope effects. Biochemistry. 1987;26:913–920. doi: 10.1021/bi00377a036. [DOI] [PubMed] [Google Scholar]
- 42.Mentch F., Parkin D.W., Schramm V.L. Transition-state structures for N-glycoside hydrolysis of AMP by acid and by AMP nucleosidase in the presence and absence of allosteric activator. Biochemistry. 1987;26:921–930. doi: 10.1021/bi00377a037. [DOI] [PubMed] [Google Scholar]
- 43.Horenstein B.A., Parkin D.W., Estupinan B., Schramm V.L. Transition-state analysis of nucleoside hydrolase from Crithidia fasciculata. Biochemistry. 1991;30:10788–10795. doi: 10.1021/bi00108a026. [DOI] [PubMed] [Google Scholar]
- 44.Admiraal S.J., O'Brien P.J. DNA-N-Glycosylases process novel O-Glycosidic sites in DNA. Biochemistry. 2013;52:4066–4074. doi: 10.1021/bi400218j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davies G.J., Gloster T.M., Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 2005;15:637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Vocadlo D.J., Davies G.J. Mechanistic Insights into glycosidase chemistry. Curr. Opin. Chem. Biol. 2008;12:539–555. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Zechel D.L., Withers S.G. Dissection of nucleophilic and acid-base catalysis in glycosidases. Curr. Opin. Chem. Biol. 2001;5:643–649. doi: 10.1016/s1367-5931(01)00260-5. [DOI] [PubMed] [Google Scholar]
- 48.Pascal M., Schuber F. The stereochemistry of calf spleen NAD-glycohydrolase-catalyzed NAD methanolysis. FEBS Lett. 1976;66:107–109. doi: 10.1016/0014-5793(76)80596-0. [DOI] [PubMed] [Google Scholar]
- 49.Dallosso A.R., Dolwani S., Jones N., Jones S., Colley J., Maynard J., Idziaszcyk S., Humphreys V., Arnold J., Donalson A., et al. Inherited predisposition to colorectal adenomas caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1 or NEIL1,2 or 3. Gut. 2008;57:1252–1255. doi: 10.1136/gut.2007.145748. [DOI] [PubMed] [Google Scholar]
- 50.Schramm V.L. Transition states, analogues, and drug development. ACS Chem. Biol. 2013;8:71–81. doi: 10.1021/cb300631k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korycka A., Blonski J.Z., Robak T. Forodesine (BCX-1777, Immucillin H)–a new purine nucleoside analogue: mechanism of action and potential clinical application. Mini Rev. Med. Chem. 2007;7:976–983. doi: 10.2174/138955707781662636. [DOI] [PubMed] [Google Scholar]
- 52.Helleday T., Petermann E., Lundin C., Hodgson B., Sharma R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 53.Jarvis L.M. Pushing cancer over the edge. Chem. Eng. News. 2013;91:13–18. [Google Scholar]
- 54.Gad H., Koolmeister T., Jemth A., Eshtad S., Jacques S., Ström C., Svensson L., Schultz N., Lundbäck T., Einarsdottir B., et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 55.Huber K., Salah E., Radic B., Gridling M., Elkins J., Stukalov A., Jemth A., Göktürk C., Sanjiv K., Strömberg K., et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature. 2014;508:222–227. doi: 10.1038/nature13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.