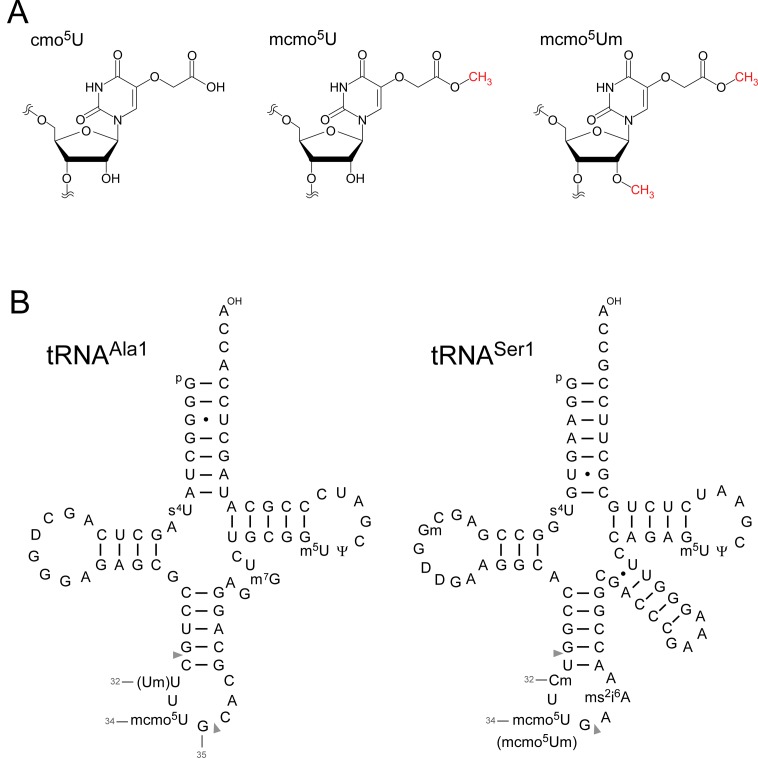
Figure 1.
5-carboxymethoxyuridine (cmo5U) and E. coli tRNAs. (A) Chemical structures of 5-carboxymethoxyuridine (cmo5U, left), 5-methoxycarbonylmethoxyuridine (mcmo5U, center) and 5-methoxycarbonylmethoxy-2′-O-methyluridine (mcmo5Um, right). (B) Secondary structures of E. coli tRNAAla1 (left) and tRNASer1 (right) with post-transcriptional modifications: 4-thiouridine (s4U), 2′-O-methylguanosine (Gm), dihydrouridine (D), 2′-O-methylcytidine (Cm), 2′-O-methyluridine (Um), 5-methoxycarbonylmethoxyuridine (mcmo5U), 5-methoxycarbonylmethoxy-2′-O-methyluridine (mcmo5Um), 2-methylthio-N6-isopentenyladenosine (ms2i6A), 7-methylguanosine (m7G), 5-methyluridine (m5U) and pseudouridine (Ψ). The position numbers of the residues (gray letters) are displayed according to the nucleotide numbering system (64). Pairs of gray triangles indicate the positions of cleavage by RNase T1 that generate RNA fragments containing the wobble positions. The sequence of E. coli tRNAAla1 is the same as that of tRNAAla1B (65).