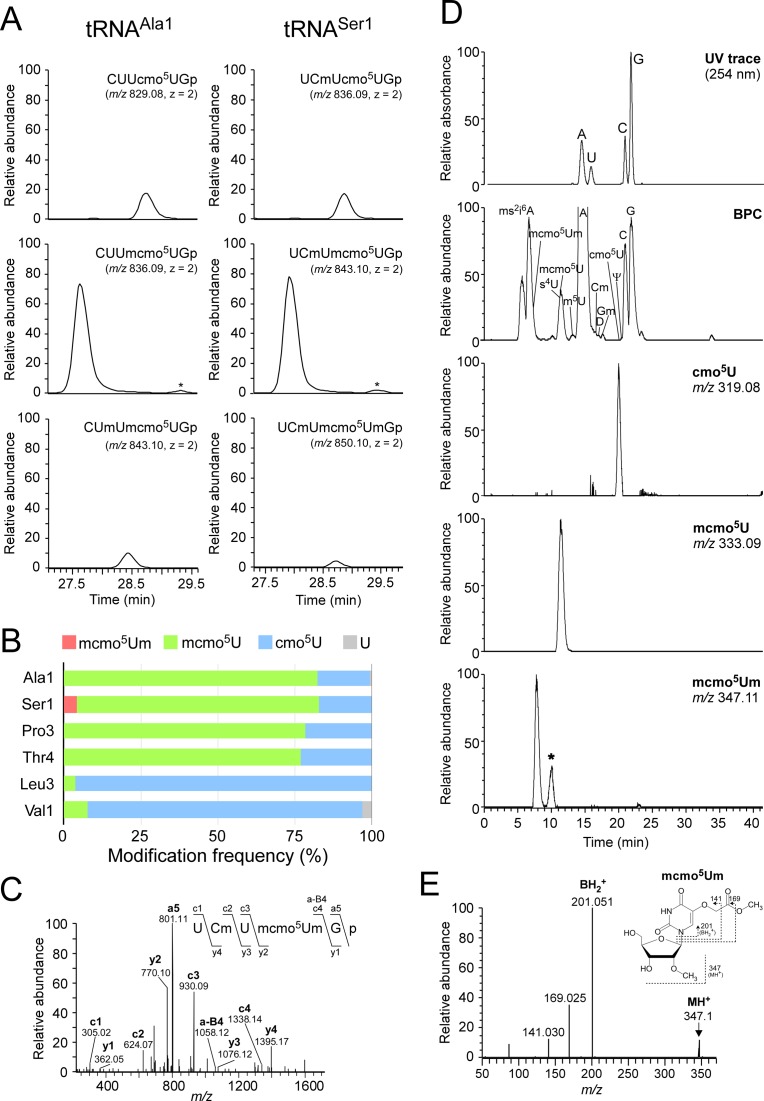
Figure 2.
Mass spectrometric analysis of individual tRNAs isolated from stationary-phase E. coli. (A) Mass chromatograms of RNase T1-digested fragments containing cmo5U and its derivatives from tRNAAla1 (left panels) and tRNASer1 (right panels) isolated from stationary-phase E. coli. Top and middle panels: extracted-ion chromatograms (XIC) for doubly-charged negative ions of cmo5U34-containing fragments (CUUcmo5UGp of tRNAAla1, MW 1660.18, m/z 829.08; UCmUcmo5UGp of tRNASer1, MW 1674.19, m/z 836.09) and mcmo5U34-containing fragments (CUUmcmo5UGp of tRNAAla1, MW 1674.19, m/z 836.09; UCmUmcmo5UGp of tRNASer1, MW 1688.21, m/z 843.10), respectively. Bottom left and bottom right panels: XICs for doubly-charged negative ions of Um32/mcmo5U34-containing fragment of tRNAAla1 (CUmUmcmo5UGp, MW 1688.21, m/z 843.10) and mcmo5Um34-containing fragment of tRNASer1 (UCmUmcmo5UmGp, MW 1702.22, m/z 850.10), respectively. The peaks marked with asterisks represent Um32/cmo5U-containing fragment (CUmUcmo5UGp, MW 1674.19, m/z 836.09) in tRNAAla1 and cmo5Um-containing fragment (UCmUcmo5UmGp, MW 1688.21, m/z 843.10) in tRNASer1. The RNA fragments containing unmodified C32 are also present in tRNASer1, but they are not described here due to high frequency of Cm32 in tRNASer1 isolated from stationary-phase E. coli. (B) Modification frequencies of cmo5U and its derivatives at position 34 in six species of tRNAs isolated from stationary-phase E. coli. Relative composition of each modification was calculated from the peak area ratio of mass chromatograms of RNase T1-digested fragments containing mcmo5Um34 (red), mcmo5U34 (green), cmo5U34 (blue) or U (gray). (C) Collision-induced dissociation (CID) spectrum of a fragment of E. coli tRNASer1 containing mcmo5Um34. The doubly-charged negative ion of the RNase T1-digested fragment containing mcmo5Um34 (m/z 850.10) was used as the precursor ion for CID. The product ions were assigned as described previously (66). Sequences of parent ion and assigned product ions are described on the upper left side of this panel. (D) Nucleoside analysis of E. coli tRNASer1. Top panel: UV chromatogram at 254 nm of the four major nucleosides (C, U, G and A). Second panel: mass chromatograms of the protonated nucleosides (MH+) on the base peak chromatogram (BPC). Third to bottom panels: XICs for cmo5U (m/z 319.08), mcmo5U (m/z 333.09) and mcmo5Um (m/z 347.11), respectively. The peak marked with an asterisk represents unspecific peak. (E) CID spectrum of mcmo5Um nucleoside. The protonated mcmo5Um (MH+, m/z 347.11) was used as the precursor ion for CID. The N-glycoside bond cleaved to generate the base-related ion (BH2+) and other product ions are assigned on the chemical structure.