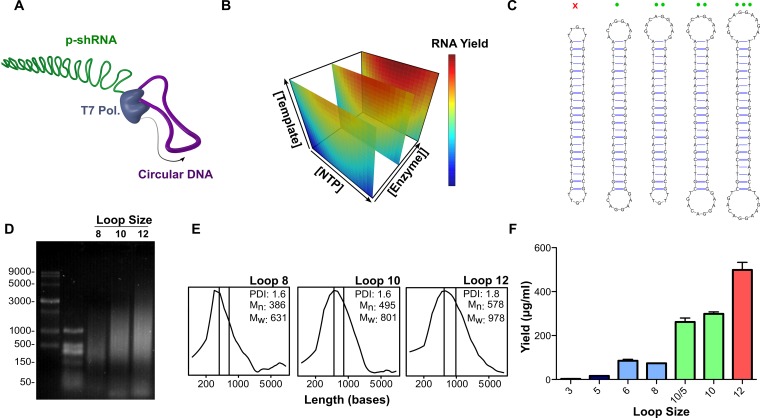
Figure 1.
Optimization of rolling circle transcription (RCT) reaction. (A) Schematic representation of RCT from a dumbbell-shaped circular DNA template. (B) Response surface model of RNA yield from RCT as a function of template, NTP and T7 RNA polymerase concentrations. RNA yield increased linearly as a function of template and polymerase concentrations, but showed a second order response to NTP concentration, with a maximum yield at ≈7 mM total NTPs. (C) RCT was carried out from a series of DNA templates with variable loop sizes. The red x indicates that no RCT occurred from the template with two 5-base loops; green circles indicate RCT yield increased as a function of loop size for loops >5 bases. (D) Denaturing agarose gel electrophoresis (1.5%, MOPS, 7% formaldehyde) of RCT reaction products obtained from the templates with 8-base, 10-base, and 12-base loops (templates 4, 6 and 11). (E) Size distributions with corresponding number avg. molecular weight (Mn), weight avg. molecular weight (Mw) and PDI, calculated from the gel in (D). (F) Quant-iT Ribogreen quantification of the RNA yield from the different dumbbell templates. Yields are reported as the mean with s.d. for two independent reactions.