Figure 3.
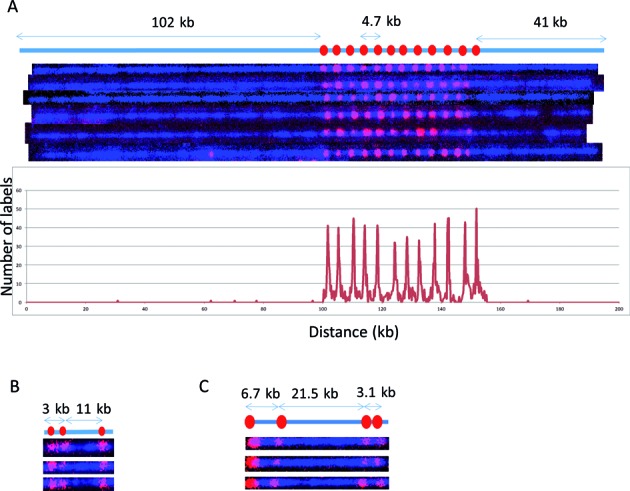
Cas9n-nick labeling. The expected labeling pattern and distances between labels is shown above corresponding fluorescent microscopy images. (A) A single copy of HLS DUF1220 triplet is approximately 4.7 kb, which cannot be identified with the sequence-motif labeling method due to lack of recognition sites within the repeat region. The Cas9n fluorescent nick-labeling method was able to label this region using a gRNA designed to the DUF1220 triplet. These recognition sites are labeled with red labels. The tandem repeats are separated approximately 4.7 kb as predicted. The histogram of the DUF1220 gRNA labels is shown below the molecules. A total of 192 molecules were evaluated for the labeling efficiency. (B) Three sgRNAs were designed to target three different locations (Gag, Pol, Env) within the HIV-1 genome. The sites were correctly labeled with the expected distances between each sgRNA. The image shows the linearized DNA after EcoRI digestion. (C) Three gRNAs designed for the chromosome 1q subtelomere generated the expected pattern. A fourth gRNA, targeting the repetitive telomere sequence (TTAGGG)n, correctly labeled the telomere region on the left end of this telomere-terminal 1q DNA fragment.
