Figure 4.
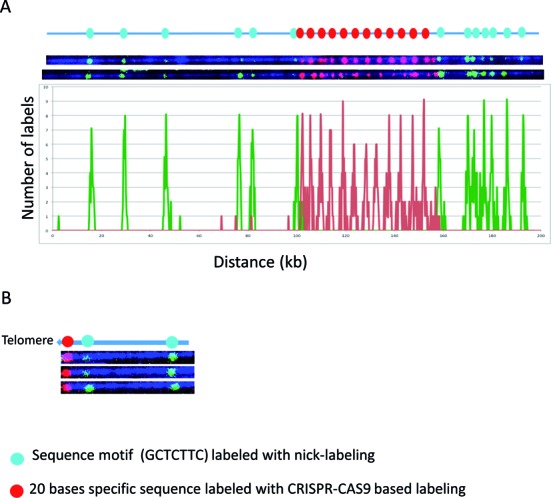
The combination of Cas9n sequence specific and nicking motif labeling. The Cas9n fluorescent nick-labeling system is effective at labeling specific repetitive sequences and locating them relative to flanking large DNA segments labeled using conventional sequence-motif labeling. The expected labeling pattern and distances between labels is shown above corresponding fluorescent microscopy images. (A) The Cas9n fluorescent nick-labeling method was used to label the repetitive sequences the DUF1220 triplet and is shown with red labels. These molecules were then labeled with the conventional sequence-motif labeling method with Nt.BspQI (green labels). The histogram of the labels is shown below the molecules. The red peaks represent the DUF gRNA labels and the green the Nt.BspQI labeling sites. A total of 43 molecules were evaluated for the labeling efficiency. (B) The Cas9n fluorescent nick-labeling method was used to label the repetitive telomere sequences at the left end of these telomere-terminal DNA fragments (red labels). The sequence-motif labeling method with Nt.BspQI was performed after Cas9n/gRNA system labeling of telomeres, and is displayed with green labels.
