Figure 4.
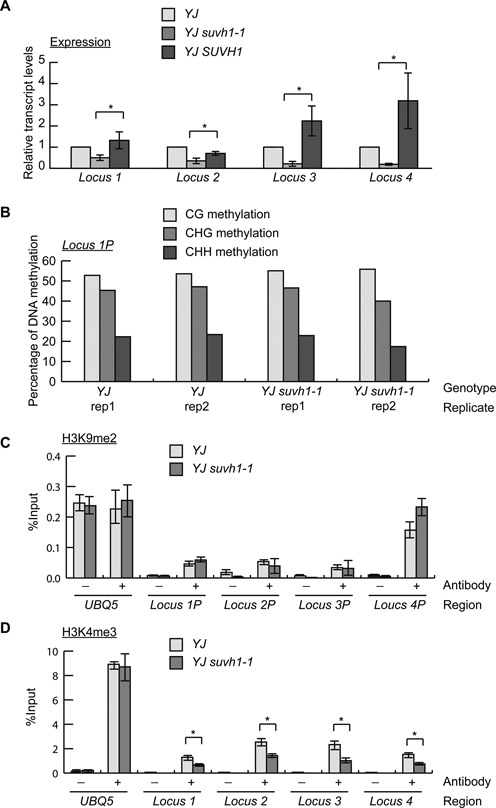
The suvh1–1 mutation leads to the reduced expression of endogenous loci with corresponding reductions in H3K4me3 levels. (A) The expression of four SUVH1-targeted endogenous loci was confirmed by RT-qPCR, and the decreased expression observed in YJ suvh1–1 was rescued in YJ SUVH1 (YJ SUVH1p:SUVH1–3XFLAG suvh1–1) for all four loci. Error bars represent standard deviations calculated from three biological replicates. * Significant difference with a P-value <0.05. (B) The DNA methylation levels of the 1 kb promoter of locus 1 determined from the two biological replicates (rep) of the YJ and YJ suvh1–1 methylome data. In all four MethylC-seq libraries, CG, CHG and CHH methylation was detected, and there were no consistent differences between YJ and YJ suvh1–1. Locus 1P represents the promoter of locus 1. (C) ChIP-qPCR was performed to measure H3K9me2 levels at the promoters of loci 1–4 in YJ and YJ suvh1–1. Locus 1P represents the promoter of locus 1, the same terminology also applies to other loci. No changes in H3K9me2 levels were observed. (D). ChIP-qPCR was performed to measure H3K4me3 levels in the coding (or transcript) regions of the four loci. Reduced H3K4me3 levels were observed in YJ suvh1–1. * Significant difference with a P-value <0.05. UBQ5 was included as a control in (C and D). Error bars representing standard deviations were calculated from three technical replicates in (C and D). Three biological replicates gave similar results. ‘–’ represents the samples without antibody; ‘+’ represents the samples with H3K9me2 or H3K4me3 antibodies added.
