Figure 1.
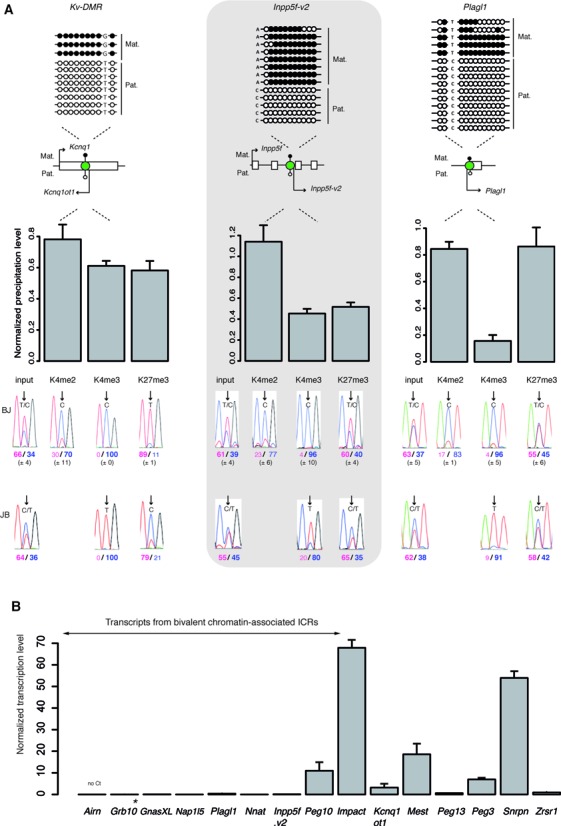
Bivalent chromatin marks the unmethylated allele of a subset of maternal ICRs in ES cells. (A) Schematic representation of the analyzed ICRs (green circles) and their position relative to the main associated gene(s). The methylation patterns are symbolized by lollipops (black: methylated; white: unmethylated). The name of the transcripts initiating from the ICR region are also indicated. Kv-DMR is an example of an ICR with a canonical chromatin signature. The Inpp5f-v2 and Plagl1 DMRs are examples of ICRs marked by mono-allelic bivalent chromatin. The upper panels show representative bisulfite-based sequencing data obtained from BJ1 ES cells. Each horizontal row of circles represents the CpG dinucleotides on an individual chromosome. Solid circles, methylated CpG dinucleotides; open circles, unmethylated CpG dinucleotides. The parental origin (Mat., maternal; Pat., paternal) was determined using the indicated strain-specific SNPs. The lower panels show the data obtained by native ChIP using anti-H3-K4me2, -K4me3 and -K27me3 antibodies. These three histone marks were similarly enriched at the analyzed ICRs in BJ1 (n = 5) and JB1 (n = 1) ES cells. The precipitation level was normalized to that obtained at the Rpl30 promoter (for H3K4me2 and H3K4me3) and at the Hoxa3 promoter (for H3K27me3). The allelic distribution of each histone mark was determined by direct sequencing of the PCR product encompassing a strain-specific SNP in the analyzed region. The mean values (± standard deviation) of the relative allelic ratios (Pink: maternal; Blue: paternal) are shown under representative chromatograms. H3K27me3 is enriched only on the maternal allele at Kv-DMR and on both alleles at the Inpp5f-v2 and Plagl1 ICRs, forming thus, with H3K4me2 and H3K4me3 a bivalent chromatin structure on the paternal unmethylated allele. (B) Microfluidic-based quantitative RT–PCR analysis of the transcripts initiating from the 15 studied maternal ICRs in BJ1 ES cells. Results are presented as the percentage of the geometrical mean of the expression of the three housekeeping genes Arbp, Gapdh and Tbp. Data were obtained from two independent experiments, each done in duplicate. *Grb10 pat-isoform.
