Figure 5.
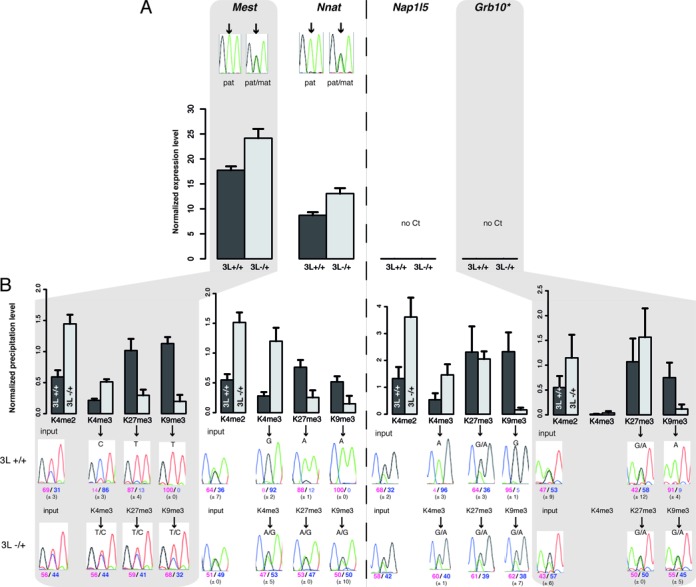
Chromatin bivalency is present at both parental alleles of transcriptionally inactive mat-ICRs in Dnmt3L−/+ embryos. Details of the molecular analyses at ICRs in WT and Dnmt3l−/+ E9.5 embryos. Mest and Nnat are representative examples of transcriptionally active and Nap1L5 and Grb10 of transcriptionally inactive ICRs in WT embryos. (A) Transcriptional analysis using pools of at least 15 WT or mutant embryos. Results are presented as the percentage of expression relative to the geometrical mean of the expression of the two housekeeping genes Ppia and Rpl30. Data are from two independent experiments, each analyzed in duplicate. No expression was detected from the Nap1l5 and Grb10 ICRs (no Ct) in both WT and mutant conceptuses. The parental origin of expression from the Mest and Nnat ICRs was determined by direct sequencing of the PCR product encompassing a strain-specific SNP in the analyzed region (* Grb10 pat-isoform). (B) Chromatin analysis following native ChIP with anti-H3-K4me2, -K4me3, -K27me3 and -K9me3 antibodies. The precipitation level was normalized to that obtained at the Rpl30 promoter (for H3K4me2 and H3K4me3), IAP (for H3K9me3) and the Hoxa3 promoter (for H3K27me3). The allelic distribution of these marks was determined by direct sequencing of the PCR product encompassing a strain-specific SNP in the analyzed region. The mean values (± standard deviation) of the relative allelic ratios (pink: maternal; blue: paternal) are indicated under representative chromatograms. Values are the mean of three independent ChIP experiments, each analyzed in duplicate.
