Figure 7.
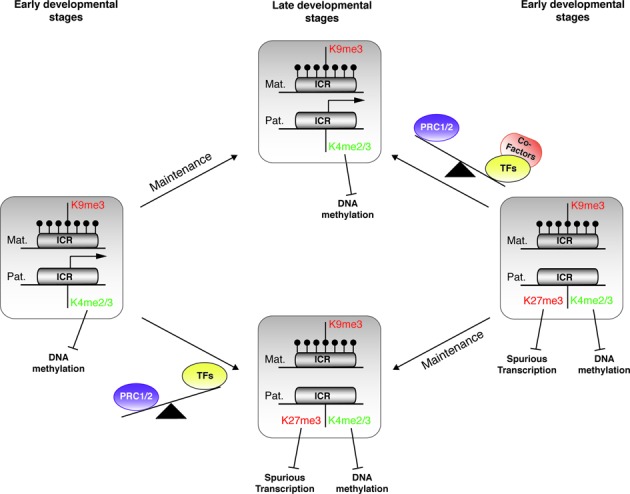
Model of the fine-tuned regulation of imprinted gene expression by chromatin bivalency. Model illustrating the gain and loss of chromatin bivalency at maternal ICRs during development. Relative to their transcriptional activity, the ICR paternal allele can be associated (right panel) or not (left panel) with bivalent chromatin in pluripotent cells (early developmental stages). This signature will be maintained, upon differentiation, in lineage sharing the same transcriptional status (late developmental stages). During lineage commitment, in the absence of transcriptional activity or transcription below a certain threshold, H3K27me3 is deposited by default on the paternal allele, already marked by H3K4me2/3, thus leading to the formation of a bivalent domain (from left to lower central panel). This signature, by repelling DNA methylation and protecting against spurious expression, contributes to protecting the ICR integrity in non-expressing cells. Conversely, the switch from gene repression to expression, upon lineage commitment, might rely on the availability of the ad hoc transcription machinery associated with H3K27me3 removal. Specific co-factors, for instance H3K27me3 demethylases, could counteract PRC1/2 action and ensure robust transcription from the paternal allele (from right to upper central panel). DNA methylation is represented by filled lollipops. Arrows indicate active transcription.
