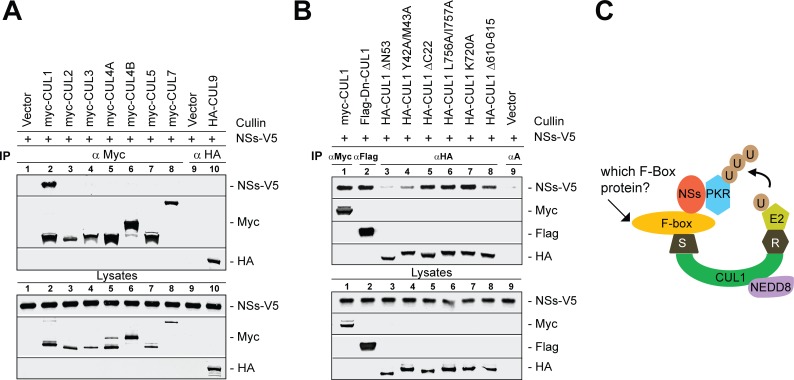
Fig 3. NSs binds to CUL1 most efficiently and regulates PKR degradation.
(A) Co-immunoprecipitation (Co-IP) of the NSs protein shows that CUL1 bound most efficiently to NSs among the 8 cullin family members, including CUL1 to 3, CUL4A-4B, CUL5, CUL7 or CUL9. Lysates of 293T cells that were transfected to express the vector alone or myc-tagged CUL1, -2, -3, -4A, -4B, -5 or -7 or HA tagged CUL9 proteins were combined with rMP-12-NSs-V5 infected cell lysates and immunoprecipitated with the anti-myc or anti-HA antibodies. The bound proteins were detected by Western blot analysis as shown in the figure. Lysate controls represent 5% of the lysate used in co-IP. (B) NSs protein binding to various CUL1 mutant proteins that were transiently expressed in 293T was determined as described above in A. αA represents IP using all three: HA, myc and Flag antibodies in the control (C) Tentative model of PKR degradation by SCF complex: CUL1 forms a molecular scaffold to organize the SCF by forming two distinct functional modules. The C terminal domain (CTD) of CUL1 (green) binds to RBX1 (R, grey), that recruits ubiquitin (U, brown) loading E2 enzyme (light green) for catalysis while the N terminus binds to SKP1 (S, grey), which recruits substrate in this case NSs (red)-PKR (blue),) via substrate recognizing F-box protein (yellow). PKR is presumably poly-ubiquitinated (UUU, brown) by the E2 enzyme and subjected to proteasome degradation. CUL1 is conjugated with NEDD8 (purple) and is essential for SCF E3 ligase activity.