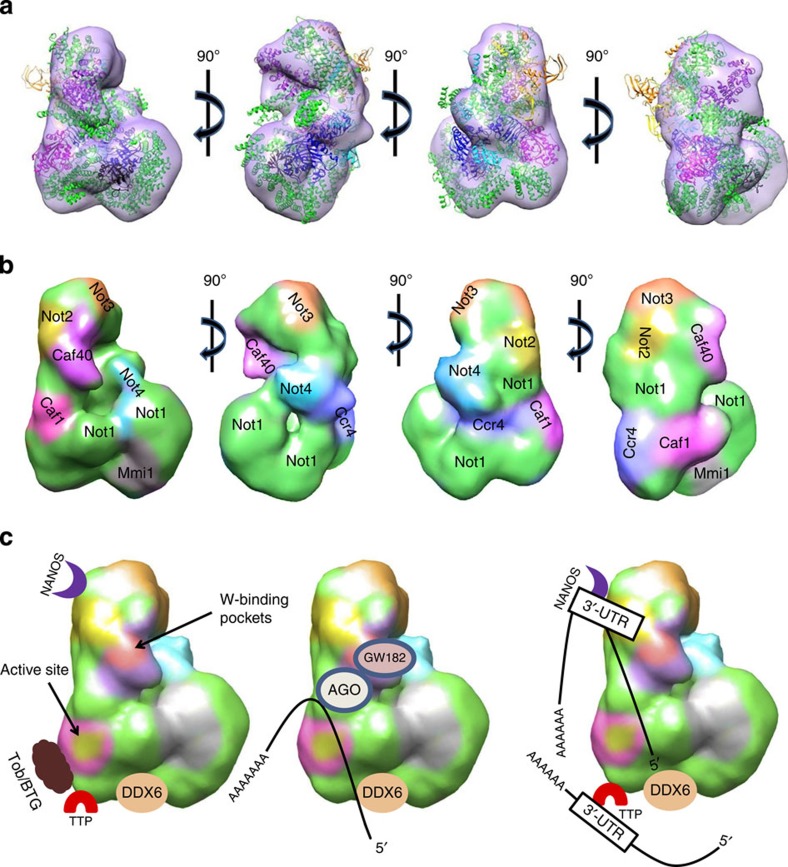
Figure 6. Functional implications of CCR4-NOT subunit organization.
(a) Pseudo-atomic model generated after model building. (highly disordered regions were removed from the model, but their location for each subunit is suggested by the color codes in b and c (b) Map of the CCR4-NOT complex subunits. Expanded coloured regions of Not2, Not3, Not4 and Mmi1 represent the disordered fragments. (c) Left, binding sites of the known CCR4-NOT-interacting partners Tob/BTG (which binds to Caf1) and Nanos, TTP and DDX6 (which bind to different parts of Not1). The Caf1 deadenylase active site is indicated in yellow and Caf40 W-binding pockets in pink. Centre, proposed mechanism of miRNA substrate degradation. Caf40 recruits RITS complex through GW182 binding; the mRNA 5′ end interacts with DDX6, which enhances its decapping activity, while the 3′ end is deadenylated by Caf1. Right, mechanism for degradation of mRNA recruited by the mRNA-binding proteins Nanos and TTP. These proteins bind to the 3′-UTR of mRNA, which they recruit to the CCR4-NOT complex.