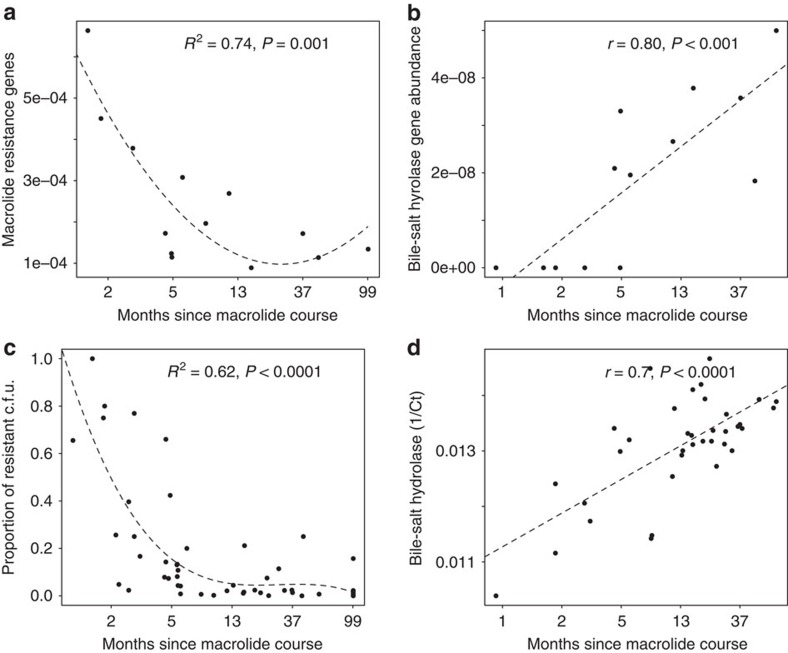
Figure 3. Macrolide resistance and bile-salt hydrolase abundance in relation to time since the last macrolide course.
The dashed lines show the model fit (linear or polynomial), R2 indicates the variation explained by the model and the P values (estimated using linear models) are indicated. (a) Macrolide resistance potential inferred from metagenomic analysis, N=14. (b) Bile-salt hydrolase abundance in metagenomes, N=14. (c) Macrolide resistance measured as proportion of anaerobic c.f.u.'s growing with erythromycin compared with c.f.u. without erythromycin, N=80. (d) Combined relative abundance of three bile-salt hydrolase genes (bsh) based on qPCR as a function of time since last macrolide course, N=37.