Abstract
Forest ecosystems need to be sustainably managed, as they are major reservoirs of biodiversity, provide important economic resources and modulate global climate. We have a poor knowledge of populations responsible for key biomass degradation processes in forest soils and the effects of forest harvesting on these populations. Here, we investigated the effects of three timber-harvesting methods, varying in the degree of organic matter removal, on putatively hemicellulolytic bacterial and fungal populations 10 or more years after harvesting and replanting. We used stable-isotope probing to identify populations that incorporated 13C from labeled hemicellulose, analyzing 13C-enriched phospholipid fatty acids, bacterial 16 S rRNA genes and fungal ITS regions. In soil microcosms, we identified 104 bacterial and 52 fungal hemicellulolytic operational taxonomic units (OTUs). Several of these OTUs are affiliated with taxa not previously reported to degrade hemicellulose, including the bacterial genera Methylibium, Pelomonas and Rhodoferax, and the fungal genera Cladosporium, Pseudeurotiaceae, Capronia, Xenopolyscytalum and Venturia. The effect of harvesting on hemicellulolytic populations was evaluated based on in situ bacterial and fungal OTUs. Harvesting treatments had significant but modest long-term effects on relative abundances of hemicellulolytic populations, which differed in strength between two ecozones and between soil layers. For soils incubated in microcosms, prior harvesting treatments did not affect the rate of incorporation of hemicellulose carbon into microbial biomass. In six ecozones across North America, distributions of the bacterial hemicellulolytic OTUs were similar, whereas distributions of fungal ones differed. Our work demonstrates that diverse taxa in soil are hemicellulolytic, many of which are differentially affected by the impact of harvesting on environmental conditions. However, the hemicellulolytic capacity of soil communities appears resilient.
Introduction
Forests remove vast amount of carbon dioxide from the atmosphere annually, equaling ~3 Pg or ~30% of fossil fuel emissions (Canadell and Raupach, 2008). Yet, deforestation results in the annual loss of forestland, the approximate size of the United Kingdom and the release of CO2 on a scale equivalent to the entire transportation sector, with accompanying loss of habitat and biodiversity (FAO, 2010). There is a need to manage forests to effectively minimize greenhouse gas emissions while maintaining sustainable production. Forest management influences the availability of soil carbon and nitrogen, which are key variables determining soil fertility (Johnson and Curtis, 2001). Soil decomposition processes mediated by microbes have a predominant role in mineralization and cycling nutrients to plants and the atmosphere by breaking down complex biomolecules (Doran, 2002; Van Der Heijden et al., 2008). Improperly managed forests can revert from net carbon sinks to net contributors of greenhouse gas emissions (Kurz et al., 2008). Improved understanding of the relationships among soils, their microbial communities and carbon cycling has great potential to contribute to forest management practices that ensure the long-term productivity while mitigating climate change.
A key question for sustainable forest management is how the removal of biomass during forest harvesting will affect carbon cycling and the long-term productivity of the secondary forest. The long-term soil productivity (LTSP) Study was established to evaluate the consequences of soft-wood lumber harvesting across multiple ecozones in North America (Powers et al., 2005). The LTSP Study has been underway for over two decades and will continue to document the long-term effects of pulse soil disturbances, particularly the removal of organic matter (OM) during harvesting. To date, short-term effects of harvesting included altered plant community, nutrient availability, soil structure, microclimate and soil litter composition (Keenan and Kimmins, 1993). However, after 10–15 years, these properties have returned to resemble pre-harvesting conditions (Powers, 1999; Sanchez et al., 2006). Generally, there were no pronounced differences among harvesting treatments with three different levels of OM removal on forest productivity (Powers et al., 2005). Relatively, little is known about the effects of these OM removal treatments on the soil microbial community. A recent study of LTSP sites in BC, Canada found that the removal of OM significantly and persistently altered both bacterial and fungal communities in soil 10–15 years following harvesting (Hartmann et al., 2012). It remains unclear if or how key functional groups within those communities, such as those involved in carbon cycling, were affected and whether such effects are broadly representative of responses in other ecozones.
Recycling photosynthetically derived carbon, roughly half of which is lignocellulose, is an essential aspect of the carbon cycle, releasing 1011 tons of monosaccharides annually. Decomposition of plant detritus provides 69–87% of nutrients required for annual forest growth (Swift et al., 1979; Sinsabaugh et al., 1993). Lignocellulosic biomass consists mainly of cellulose (35–50%), hemicellulose (25–30%) and lignin (25–30%), and is recalcitrant to chemical treatments, creating an obstacle in efficient production of lignocellulose biofuels. Hemicelluloses have an integral role stabilizing the components of lignocellulose by forming a robust network of cross-linked polymers, with hemicellulose covalently linked to lignin (Shallom and Shoham, 2003). Unlike cellulose, hemicelluloses are heterogeneous polymers consisting of furanoses, pyranoses and a variety of sugar acids, and they are branched with short lateral chains of varying saccharides. Degradation of hemicellulose is mediated by a consortium of fungi and bacteria using a variety of extra-cellular hydrolytic and oxidative enzymes (Pérez et al., 2002). Although this degradation process is understood, we have a poor knowledge of the microorganisms responsible for hemicellulose degradation in particular environments. In the context of forest management, improved knowledge of hemicellulose degraders in soil communities will advance our understanding of how disturbances affect this functional group, and more broadly, affect carbon cycling. Improved knowledge of hemicellulose degraders may also lead to the discovery of novel biocatalysts, with applications in transforming biomass to valuable chemicals and materials.
Here, we used stable-isotope probing (SIP) of DNA to identify bacterial and fungal populations from forest soil communities that assimilate carbon from hemicellulose. We further used extensive molecular surveys of microbial communities in forest soils to evaluate how these populations were impacted by timber harvesting and to determine their distributions in coniferous forests across North America. These DNA-based approaches were complemented with measures of microbial activity based on 13C incorporation into phospholipid fatty acids. This is the first study to combine these approaches to investigate both bacterial and fungal hemicellulose degraders and to investigate responses of these populations to disturbance in a large-scale field experiment.
Materials and methods
Sample collection
Samples were collected from 18 LTSP Study experimental sites in six different ecozones across North America (Table 1, Supplementary Table S1). The ecozones represent distinct climatic regimes: interior Douglas fir (IDF), cold semi-arid; sub-boreal spruce, western montane; black spruce, boreal moist; jack pine, cool, wet, moist; ponderosa pine (PP), Mediterranean; and loblolly pine, subtropical moist. Each site had controls and timber-harvesting treatment plots, representing three levels of organic matter removal: OM0, unharvested reference; OM1, stem-only harvesting (crown and branches left on site); OM2, whole-tree harvesting; and OM3, whole-tree harvesting plus removal of forest floor (organic soil layer). These harvesting treatments removed the following proportions of above-ground organic matter: OM1, 40–70% OM2, 70–90% OM3, ~100% (Powers, 2006). Plots were harvested and replanted from 10 to 15 years prior to sampling for this study. The trees used for replanting correspond to the ecozone names, except that lodgepole pine was planted in the IDF ecozone. Soils were sampled from 9 or 15 randomly selected points in each rectangular treatment plot. The organic layer and top 120 cm of the mineral layer were separately sampled. Sets of three or five corresponding samples were composited to yield triplicate samples from each layer in each treatment plot. Further details of soil sampling are described by Hartmann et al. (2012) The OM3 treatments in the IDF, sub-boreal spruce and jack pine ecozones lacked sufficient organic layers for sampling.
Table 1. LTSP Study sites sampled for this study.
| Ecozone | Location | Site | Latitude (°North) | Longitude (°West) | Elevation (m) | Annual precipitation (cm) | Precipitation, warmest quarter (mm) | Mean annual temperature (°C) |
|---|---|---|---|---|---|---|---|---|
| Dairy Creek | 50.51 | 102.25 | 1075 | |||||
| IDF | British Columbia | O'Connor Lake | 50.53 | 120.21 | 1180 | 30–75 | NA | 1.6–9.5 |
| Black Pines | 50.56 | 120.17 | 1150 | |||||
| Log Lake | 54.35 | 122.61 | 785 | 62 | ||||
| SBS | British Columbia | Skulow Lake | 52.32 | 121.92 | 1050 | 43 | 146–193 | 2.2 |
| Topley | 52.32 | 126.31 | 1100 | 53 | ||||
| Blodgett | 38.88 | 120.64 | 1320 | 165 | ||||
| PP | California | Brandy City | 39.55 | 121.04 | 1130 | 190 | 51–55 | 11.2 |
| Lowell Hill | 39.26 | 120.78 | 1270 | 173 | ||||
| Fensom 1 | 89.41 | 49.07 | 442 | |||||
| BS | Ontario | Fensom 2 | 89.38 | 49.08 | 450 | 61 | 266 | 0.4 |
| Fensom 3 | 89.39 | 49.07 | 442 | |||||
| Superior 1 | 47.58 | 82.79 | 458 | 82 | ||||
| JP | Ontario | Superior 2 | 47.58 | 82.81 | 461 | 83 | 250 | 1.7 |
| Superior 3 | 47.57 | 83.84 | 426 | 85 | ||||
| Kurth-1 | ||||||||
| LP | Texas | Kurth-2 | 31.11 | 95.15 | 88 | 109 | 253 | 19 |
| Kurth-3 |
Abbreviations: BS, black spruce; IDF, interior Douglas fir; JP, jack pine; LP, loblolly pine; NA, not available; PP, ponderosa pine; SBS, sub-boreal spruce.
SIP soil microcosm conditions
Microcosms used for SIP experiments were prepared in 30-ml serum vials (Wheaton glass) with 0.75 or 1.00 g dry weight of organic or mineral layer soil, respectively. Soil microcosms were prepared with soil samples from the IDF and PP ecozones. All microcosms were in pairs, one with 13C-labeled hemicellulose and one with unlabeled hemicellulose. In SIP-PLFA (phospholipid-derived fatty acids) experiments, one microcosm was prepared for each triplicate soil sample. In an initial SIP-DNA experiment, one microcosm was prepared for each triplicate soil sample, whereas in later experiments, triplicate soil samples were composited (Supplementary Figure S1). The moisture content was adjusted to 60% of total weight. All microcosms were pre-incubated for 7 days at 20 oC in the dark prior to the addition of substrate. Following pre-incubation, 10 mg of finely ground maize-derived hemicellulose was added, and mixed thoroughly with the soil. Hemicellulose in controls had the natural abundance of 13C (<1.2 atom % IsoLife; N-10509, Lot:0901-0238); alternatively 13C-labeled hemicellulose was used (97 atom % IsoLife; U-10509, Lot: 0901-0273). The microcosms were crimp sealed with a butyl rubber stopper and further incubated for 48 h.
SIP-PLFA
PLFAs were extracted as previously described (Bligh and Dyer, 1959; Frostegård et al., 1991) and analyzed with a gas chromatograph isotope-ratio mass spectrometer (UC Davis Stable Isotope Facility; Davis, CA, USA). The combination of C18:2n6, C18:1n9 and C18:3n3 fatty acids was used to determine fungal biomass, whereas terminal branched fatty acids i14:0, i15:0, a15:0, i16:0, i17:0 and a17:0 were used for Gram-positive bacterial biomass, and cyclopropyl and monounsaturated fatty acids cy17:0, c16:1n7 and c16:1n5 were used for Gram-negative bacterial biomass (Oravecz et al., 2004; Moore-Kucera and Dick, 2008).
Cesium chloride gradient ultracentrifugation
Total genomic DNA was extracted from soil microcosms using MPBio FastDNA Spin Kit for Soil, as recommended by the manufacturer. DNA concentrations were quantified using the PicoGreen fluorescent dye assay (Invitrogen, Carlsbad, CA, USA), and 3 μg of DNA was used for each cesium chloride gradient ultracentrifugation. DNA gradients were formed, separated into 20 fractions and purified as previously described (Neufeld et al., 2007). The enrichment of 13C-DNA was measured according to Wilhelm et al (2014). In brief, a 5-μl volume of each fraction was hydrolyzed in 100 μl of 88% formic acid at 70 oC for 1 h. Following hydrolysis, fractions were dried in a SpeedVac with an acid trap and suspended in 30 μl of 1% acetic acid (v/v). 13C levels were determined by measuring the isotopic enrichment of individual adenine and guanine nucleotides using an ultra-high-performance liquid chromatograph coupled to a tandem mass spectrometer. Fractions from each gradient containing >50% 13C DNA (average densities 1.725–1.735 g ml–1) were pooled.
Pyrotag sequencing
DNA concentrations were quantified using the PicoGreen fluorescent dye assay. Each set of pooled gradient fractions was diluted to 0.25 ng μl−1 DNA prior to PCR amplification. The bacterial V1–V3 hypervariable regions of the 16 S rRNA gene were amplified using universal 27 F and 519 R (Weisburg et al., 1991; Turner et al., 1999) barcoded bacterial primers. The fungal internal transcribed spacer (ITS2) region was amplified using barcoded primers ITS3R and ITS4F (White et al., 1990). PCR reactions were performed in triplicate for each sample, and pooled prior to purification and quantification (Hartmann et al., 2012). Samples were sequenced using the Roche 454 Titanium platform (GS FLX+) at the McGill University and Genome Québec Innovation Centre with a maximum of 40 samples multiplexed on each quarter plate, yielding an average of 4200 bacterial and 3800 fungal pyrotags per sample.
Sequence processing and analysis
Bacterial sequences were processed using the MOTHUR standard operating procedure (Schloss et al., 2011). This process entails an implementation of PyroNoise to reduce sequencing error, followed by trimming of barcoded sequencing primers, and setting a sequencing score cutoff of 350 flows. Sequences were de-replicated for subsequent steps. Chimeric sequences were filtered out using the uchime software implemented in MOTHUR. Bacterial operational taxonomic units (OTUs) were binned at a distance of 0.03, using the k-nearest neighbor algorithm (Schloss and Westcott, 2011). Fungal sequences were binned into OTUs at a Levenshtein distance of 11 using CrunchClust (Hartmann et al., 2012). To assess in situ distributions of hemicellulolytic OTUs, the bacterial and fungal sequences were clustered into OTUs with previously constructed pyrotag libraries derived from in situ soil community DNA from the six ecozones (Hartmann et al., 2012; VanInsberghe et al., 2015; Short read archive study accession (ERS713483–ERS713552)). Both bacterial and fungal OTU matrices were normalized by subsampling to 1500 sequencing reads per sample. For taxonomy classification, representative OTUs were selected by MOTHUR using the distance method, and the representatives were identified with the SILVA 16 S rRNA v. 102 reference database (Quast et al., 2013) for bacterial pyrotags and the UNITE v. 6 (Kõljalg et al., 2013) database for fungal pyrotags. The representative OTU sequences were deposited in Genbank under accession numbers KP411926 through KP412081 for uncultured bacterial and fungal clones.
Statistical analyses were performed in R and PRIMER 6 with the PERMANOVA+ add-on. All PERMANOVA analyses utilized the permutation of residuals under a reduced model with 999 permutations, using the Bray–Curtis dissimilarity index. The R Boruta package was used for random forest analysis. Species diversity and evenness were calculated using the R Vegan package (Dixon, 2003). A dendrogram was generated using Arb-Silva by a quick-parsimony addition of pyrotag sequences into a small subunit rRNA tree provided by Silva (Quast et al., 2013) and visualized using the web-based Interactive Tree of Life (www.itol.embl.de). The web-based RDP SeqMatch tool (Cole et al., 2009) was used to identify isolate strains most closely related to OTUs for use as reference sequences in the dendrogram.
Results
13C incorporation into biomass
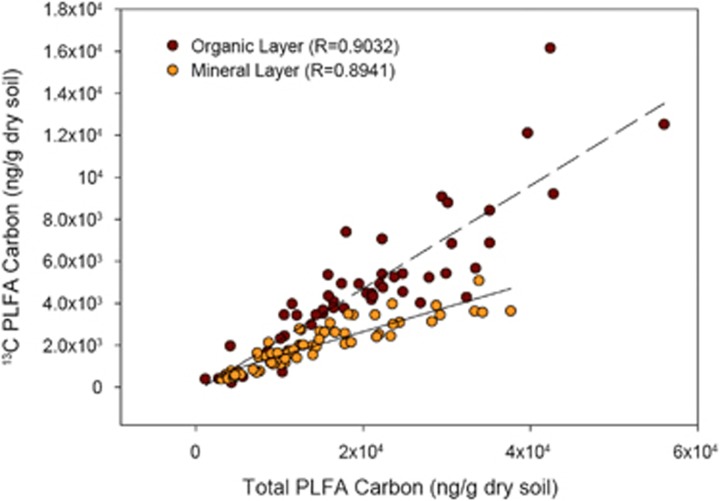
Incorporation of 13C into PLFAs and DNA was consistently observed following incubation of microcosms with 13C-hemicellulose and soils from either the IDF or PP ecozone for 48 h. Incorporation of 13C was greater in organic soil compared with mineral soil (Figure 1). There was a strong correlation between total PLFA carbon, which is proportional to total biomass, and the total amount of 13C incorporated into PLFAs, which approximates hemicellulose assimilation activity. This correlation was positive and linear, but the slope of the line differed substantially between microcosms with soil from the organic versus mineral layer. Based on domain-specific PLFAs, the relative proportion of bacterial and fungal 13C incorporation differed between soils from the IDF ecozone versus the PP ecozone, with a greater proportion of fungal incorporation in the latter (Supplementary Figures S2 & S3). There was a trend of increasing 13C incorporation into PLFAs with increasing degrees of OM removal in the forest-harvesting treatments (Supplementary Figure S4), but incorporation was highly variable and did not differ significantly among treatments.
Figure 1.
Relationship between the incorporation of hemicellulose and total biomass after microcosm incubations (n=52).
Identification of hemicellulolytic OTUs
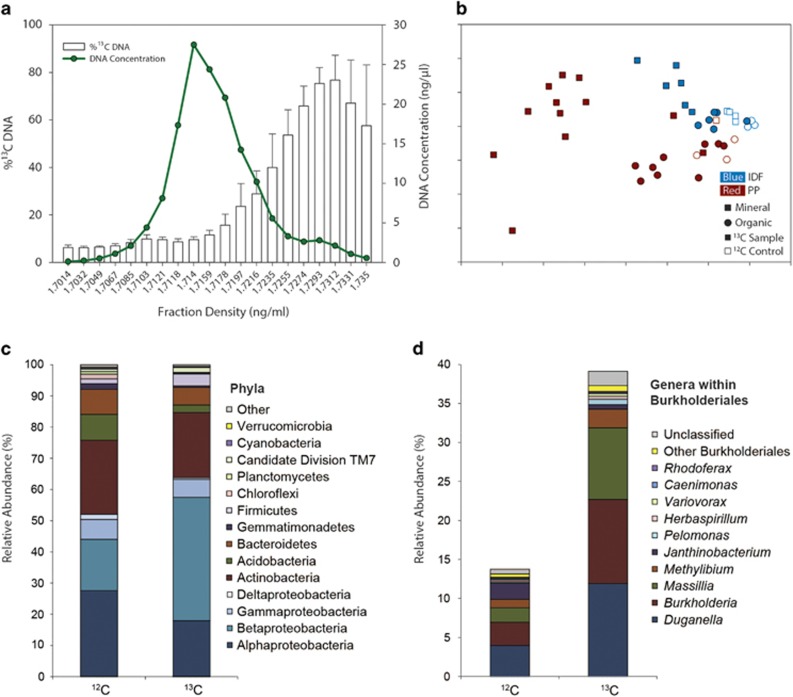
Pyrotag libraries derived from the heavy DNA fractions were compared between 13C microcosms and corresponding 12C controls to identify OTUs that assimilated carbon from hemicellulose. Substantial 13C enrichment in DNA from 13C microcosms was found in density gradient fractions between 1.725 and 1.735 g ml−1, corresponding to the first five to six fractions collected (Figure 2a). These fractions were pooled for pyrotag analysis, as were the corresponding fractions from 12C controls. However, owing to the limited amount of DNA in the heavy fractions from 12C controls, 21 of 35 bacterial control samples, and 31 of 35 fungal control samples did not yield sufficient PCR product for sequencing. To identify organisms incorporating 13C from hemicellulose, the OTU matrices from 13C microcosms and 12C controls were separately pooled and compared. This was done separately for each soil layer in each ecozone. Relative abundance profiles of bacterial and fungal OTUs differed significantly (P<0.001, PERMANOVA) between 13C and 12C microcosms (Figure 2b). OTUs with relative abundances at least fivefold greater in 13C microcosms versus the corresponding 12C controls were identified as 13C-enriched, or putatively hemicellulolytic), OTUs. A total of 104 (Supplementary Table S3) bacterial and 52 fungal (Supplementary Table S4) hemicellulolytic OTUs were identified. The greatest number of 13C-enriched bacterial OTUs were Betaproteobacteria (Figure 2c), with Burkholderia, Duganella and Massillia being the most abundant genera (Figure 2d). Of the hemicellulolytic bacterial OTU sequences, 43 were <97% identical to those of cultured strains represented in GenBank. Hemicellulolytic fungal OTUs consisted largely of Ascomycota. Of the hemicellulolytic OTUs, 20 of the bacterial ones and eight of the fungal ones were identified in soil from both ecozones. All fungal hemicellulolytic OTUs identified in the mineral layer were also present in the organic layer.
Figure 2.
An overview of analysis of pyrotags from heavy DNA fractions showing (a) separation of 13C-enriched DNA through CsCl density gradient centrifugation; (b) the non-metric multidimensional scaling ordination of pyrotag profiles from 13C microcosms (closed symbols) and 12C controls (open symbols); relative abundances in all 13C microcosms and 12C controls of phyla (c) and of genera within the order Burkholderiales (d).
Hemicellulolytic OTUs in situ
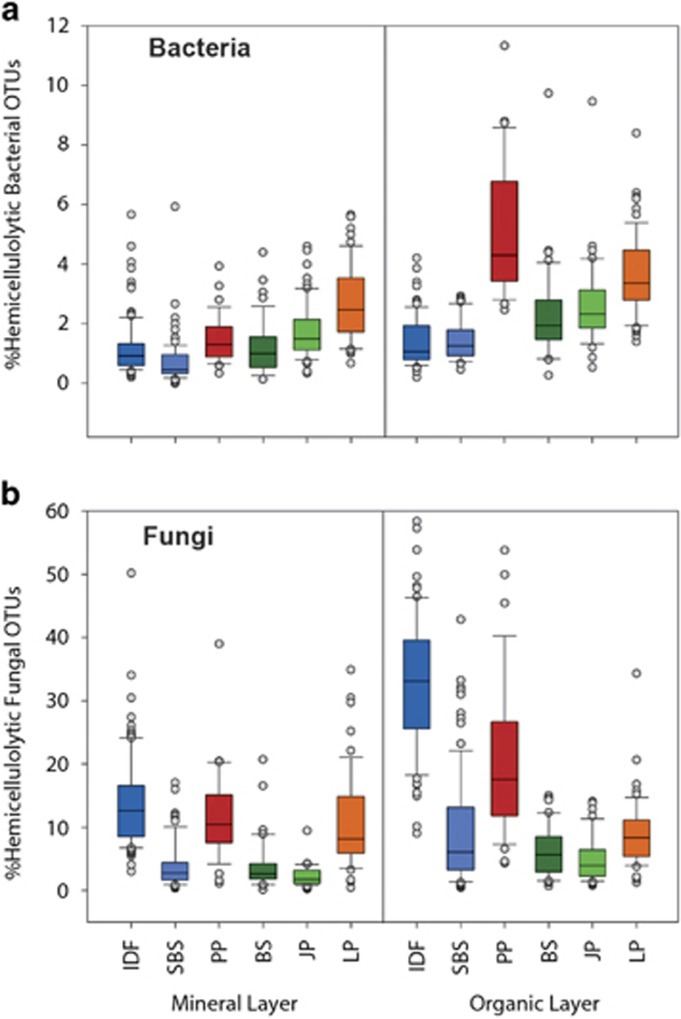
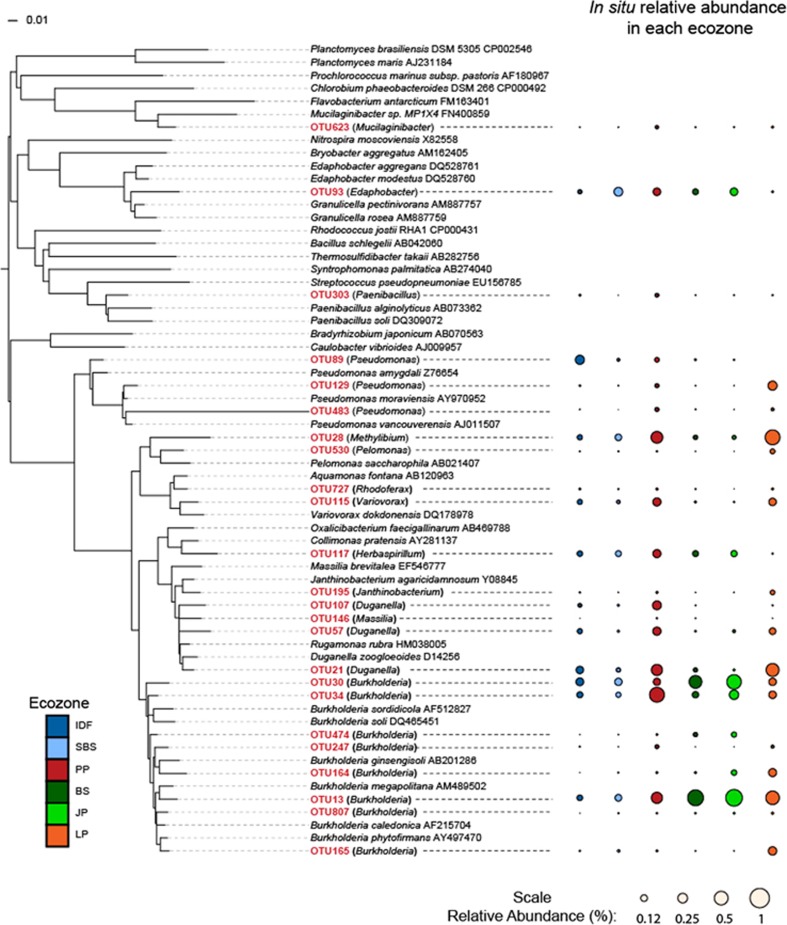
In situ relative abundances of the bacterial and fungal hemicellulolytic OTUs were analyzed within a large and highly replicated pyrotag data set representing 691 soil samples from 18 forest sites in six ecozones (Hartmann et al., 2012; VanInsberghe et al., 2015). Hemicellulolytic bacterial OTUs constituted an average of 2.0% of the total bacterial pyrotags from both organic and mineral soil layers across the six ecozones, and were most abundant (5.1%) in the organic layer of the PP ecozone (Figure 3a). Of the 24 hemicellulolytic bacterial OTUs most abundant in situ, 18 were Betaproteobacteria, including eight Burkholderia, and the remainder were Gammaproteobacteria, Firmicutes, Bacteroidetes and Acidobacteria (Figure 4). These abundant OTUs generally had similar distributions across ecozones. However, distributions of all 104 hemicellulolytic bacterial OTUs differed between the ecozones IDF and PP, and between soil layers, and PERMANOVA R2 values indicate that ecozone explains 8% of the variability in the data, whereas soil layer explains 20% (Table 2). A greater proportion of the soil community is hemicellulolytic in the organic layer than that of the mineral layer for both bacterial and fungal populations (P<0.001, Wilcoxon signed-rank test).
Figure 3.
Proportions of hemicellulolytic OTUs in in situ pyrotag libraries.
Figure 4.
Neighbor-joining tree showing the taxonomic affiliations and in situ relative abundances of the predominant hemicellulolytic bacterial OTUs (red) having >0.01% total abundance in the total pyrotag data set. Reference sequences have GenBank accession numbers.
Table 2. For selected factors, effect sizes and explained variances on in situ in relative abundances of hemicellulolytic OTUs in the IDF and PP ecozones, based on PERMANOVA.
| Factor |
Bacteria |
Fungi |
||
|---|---|---|---|---|
| F/t | R2 | F/t | R2 | |
| Ecozone | 13.88*** | 0.082 | 74.69*** | 0.368 |
| Soil Layer | 31.06*** | 0.199 | 25.01*** | 0.123 |
| Harvesting treatment | 2.38*** | 0.016 | 4.00*** | 0.027 |
| OM0 vs OM1 | 1.55** | 1.44* | ||
| OM0 vs OM2 | 2.00*** | 1.75** | ||
| OM0 vs OM3 | 2.16*** | 2.19*** | ||
| OM1 vs OM2 | 1.11ns | 1.57* | ||
| OM1 vs OM3 | 1.30ns | 2.59*** | ||
| OM2 vs OM3 | 0.86ns | 2.06*** | ||
| Ec. × layer | 13.18*** | 0.161 | 5.94*** | 0.050 |
| Ec. × treatment | 1.83** | 0.019 | 1.93** | 0.017 |
| Layer × treatment | 1.89*** | 0.022 | 2.70*** | 0.033 |
| Ec. × layer × treatment | 1.19ns | 0.007 | 1.55 ns | 0.017 |
Abbreviations: Ec., ecozone; ns, not significant. *P<0.05, **P<0.01, and ***P<0.001.
Test statistics include pseudo F-ratio (F) for main perMANOVA test, univariate t-statistic (t) for pairwise tests, and estimation of variance components (R2).
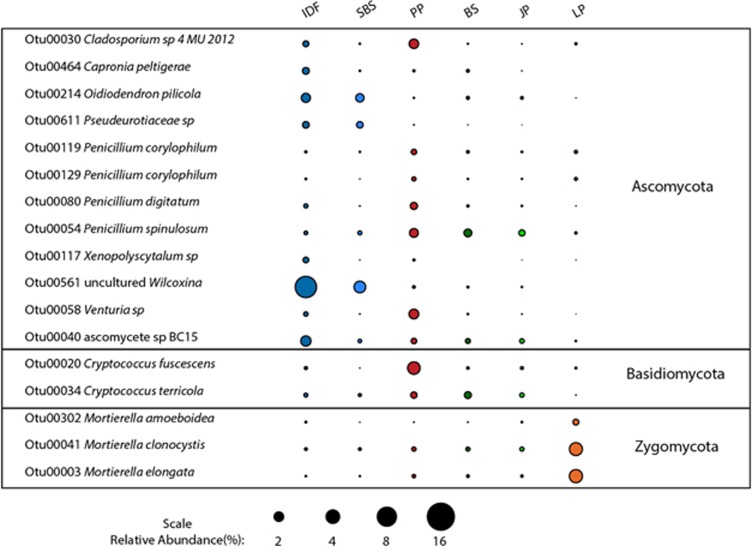
Hemicellulolytic fungal OTUs were more unevenly distributed across ecozones than bacterial ones. The fungal OTUs constituted an average of 20% of the total fungal pyrotags from both organic and mineral soil layers between IDF and PP, but this value ranged from a high of 33% from the organic layer of the IDF ecozone to a low of 12% from the mineral layer of the PP ecozone (Figure 3b). Even those hemicellulolytic fungal OTUs most abundant overall were of low abundance or absent in several ecozones (Figure 5). With some exceptions, the greatest abundances occurred in the IDF and PP ecozones whose soil was used in the microcosms to identify the hemicellulolytic OTUs. In accordance with their patchy distributions, relative abundance profiles of hemicellulolytic fungal OTUs clustered strongly according to ecozones, and PERMANOVA indicates that ecozone explains 37% of the variability in the fungal OTU abundances, far more than explained by soil layer (Table 2).
Figure 5.
In situ relative abundances of hemicellulolytic fungal OTUs having >0.01% relative abundance in the total pyrotag data set.
Bacterial hemicellulolytic populations overlapped greatly between ecozones, whereas fungal ones did not. The relative abundances of hemicellulolytic bacterial OTUs in the IDF and PP ecozones correlated strongly with each other in both the organic and mineral layers (R=0.81 and 0.66, respectively). By contrast, the relative abundances of putatively hemicellulolytic fungal OTUs in those ecozones did not correlate well (R=0.06 and 0.07 in the organic and mineral layers, respectively).
Harvesting impacts on hemicellulolytic OTUs
Timber-harvesting treatments had relatively subtle effects on distributions of hemicellulolytic OTUs that varied among the IDF and PP ecozones. Harvesting treatments did not significantly affect the total abundance of hemicellulolytic OTUs (as a proportion of total pyrotags), either in the combined data set or when each soil layer of each ecozone was separately analyzed. And, harvesting treatments had no detectable effect on the evenness of hemicellulolytic OTUs. However, harvesting treatments had a relatively small but statistically significant impact on relative abundance profiles of bacterial and fungal hemicellulolytic OTUs. For both bacterial and fungal OTUs, canonical analysis of principal coordinates ordination revealed distinct clustering of the unharvested reference (OM0) and whole-tree harvesting plus forest floor removal (OM3) treatments, whereas stem-only (OM1) and whole-tree (OM2) harvesting treatments formed a third cluster (Figure 6). Consistent with this clustering, most misclassifications in the canonical analysis of principal coordinates analysis of bacterial populations occurred in the OM1 and OM2 treatments, indicating stronger separation strengths for the OM0 and OM3. In pairwise PERMANOVA comparisons, only bacterial profiles in OM1 and OM2 treatments were not significantly different (Table 2). Fungal profiles in OM1 and OM2 treatments were significantly different, but the effect size, based on the univariate t-statistic, is smaller than those in all other pairwise comparisons of fungal profiles. When profiles for each soil layer in each ecozone were separately examined, the magnitude of harvesting treatment effects on OTU profiles differed greatly, and those effects were not universally significant (Supplementary Figure S5,Supplementary Table S2). Overall, treatment effects tended to be greater for fungal versus bacterial profiles.
Figure 6.
Canonical analysis of principal coordinates ordination based on Bray–Curtis dissimilarity of profiles of hemicellulolytic OTUs within pyrotag libraries from both soil layers of the IDF and PP ecozones. Differences among samples were maximized according to harvesting treatments. Classification success rates for each harvesting treatment are given in parentheses and the canonical correlation of each axis is given in parentheses.
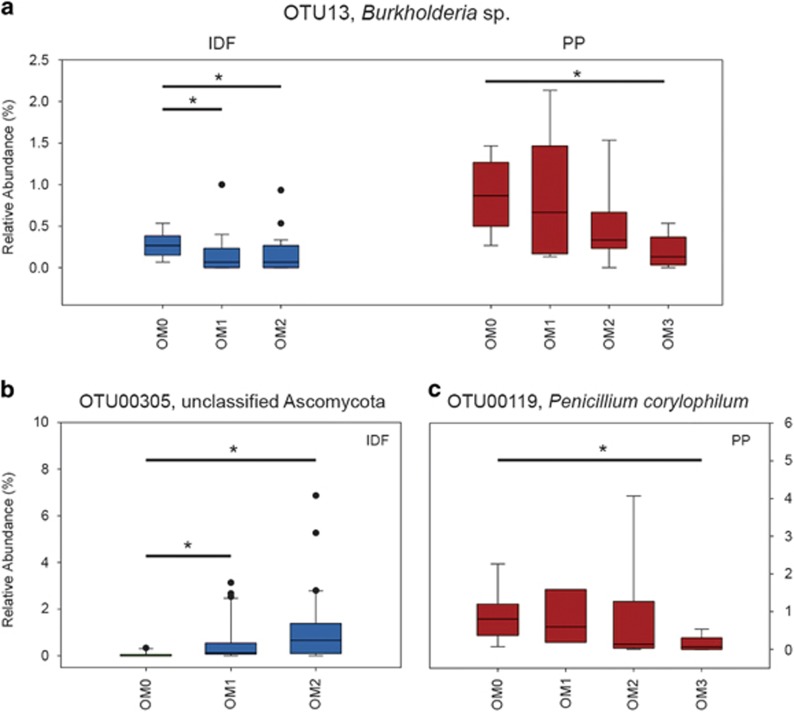
Relative abundances of most individual hemicellulolytic OTUs were not consistently affected by harvesting treatments on the basis of the Kruskal–Wallis test (with multiple comparisons). This was the case when the entire data set was analyzed and when each soil layer in each ecozone was separately analyzed. There were notable exceptions, including the hemicellulolytic OTU most abundant overall, OTU13 (Burkholderia sp.), whose abundance decreased in response to increasing organic matter removal in three ecozones (Figure 7). As well, two of the most abundant fungal OTUs, OTU00305 (Unclassified Ascomycota) and OTU00119 (Penicillium corylophilum), were affected by harvesting, each in a single ecozone.
Figure 7.
Effects of harvesting treatments on relative abundances of major hemicellulolytic OTUs in the IDF and PP ecozones (*bars denote significant pairwise differences, P<0.05).
Discussion
This investigation identified 104 bacterial and 52 fungal hemicellulolytic populations in coniferous forest soil communities from two very different ecozones in North America. These populations are estimated to account for 2% of the bacterial and 10–30% of the fungal communities in situ and likely have important roles in lignocellulose decomposition and carbon cycling. Several lines of evidence support the reliability of the SIP method in identification of these populations. The populations identified were OTUs from highly 13C-enriched DNA and were absent or much less relatively abundant in DNA of the same density from control microcosms with 12C hemicellulose. The direct measurement of 13C-enriched adenine and guanine nucleotides by ultra-high performance liquid chromatography-tandem mass spectrometry with multiple-reaction monitoring (Wilhelm et al., 2014) was invaluable in efficiently optimizing the microcosm experiments and ensuring high 13C enrichment of DNA prior to sequence analysis. The strict criteria for identifying 13C-enriched OTUs minimized false positives; however, as a consequence, some hemicellulose degraders may have been overlooked if (i) they grew very slowly or not at all in the microcosms, (ii) they were of low relative abundance or (iii) they have high GC content causing their DNA to occur in the heavy fractions of the 12C control DNA. The short, 48-h, incubations limited the movement of label beyond the primary hemicellulose consumers via biomass turnover. However, we cannot exclude the possibility that we identified some ‘cheaters' that assimilated hydrolysis products of hemicellulose without producing extracytoplasmic enzymes necessary to hydrolyze hemicellulose. The above caveats are unavoidable in SIP experiments. The correct identification of hemicellulose-degrading populations was directly confirmed in two cases, OTU30 and OTU1367 whose pyrotag sequences match those of strains isolated on xylan, a major constituent of hemicellulose (R. Wilhelm, unpublished data). Finally, many of the hemicellulose-assimilating OTUs belong to taxa previously reported to grow on hemicellulose, as described below.
The hemicellulolytic bacterial populations identified represent diverse taxa from four phyla (Figure 4). However, those populations most abundant in situ were mainly Proteobacteria. These include members of Burkholderia and Pseudomonas, two genera known to degrade a wide variety of organic compounds and previously implicated in lignocellulose degradation (Harazono et al., 2003; Mohana et al., 2008; Cheng and Chang, 2011). Both genera occupy diverse ecological niches, as soil and aquatic heterotrophs, plant mutualists as well as plant and animal opportunistic pathogens (Coenye and Vandamme, 2003; Salles et al., 2004). The hemicellulolytic Burkholderia identified here are affiliated with plant-associated saprophytes belonging to the Group A Burkholderia lineage (Estrada-de los Santos et al., 2013) as determined by RDP SeqMatch (Cole et al., 2009). Additional populations identified belong to Duganella and Variovorax as well as the non-Proteobacteria, associated with Mucilaginibacter (Bacteroidetes) and Paenibacillus (Firmicutes). All four of these genera were previously reported to be involved in hemicellulose degradation (Ghio et al., 2012; Maki et al., 2012; Štursová et al., 2012; Talia et al., 2012; Khan et al., 2013a, b). Another population identified is associated with Granulicella, a genus within the Acidobacteria, which is abundant in soil but poorly characterized because of a lack of cultured representatives. Members of Granulicella have been reported to grow on xylan (Pankratov and Dedysh, 2010). Other members of the Acidobacteria are reported to grow on hemicellulose (de Castro et al., 2013), and genomic analysis has supported the proposition that polysaccharides are important substrates for Acidobacteria (Ward et al., 2009; Rawat et al., 2014). This is the first report of hemicellulolytic activity in Betaproteobacteria belonging to the genera, Methylibium, Pelomonas and Rhodoferax, Methylibium, which was abundant in all ecozones (Figure 4), was previously known to include facultative methylotrophs and able to utilize one-carbon substrates and aromatic compounds (Nakatsu et al., 2006; Song and Cho, 2007). Genomes of members of three of these genera, M. petroleiphilum and R. ferrireducens and Pelomas sp., reveal several enzymes that are predicted to act on hemicellulose (Supplementary Table S5).
The hemicellulolytic fungal populations identified also represent diverse taxa from three fungal phyla (Figure 5). These populations include members of Mortierella and Cryptococcus, genera previously known to be involved in hemicellulose degradation (Biely et al., 1978; Morosoli et al., 1993; Chávez et al., 2006; Varnaitė and Raudonienė, 2008; Zeng et al., 2013). A third genus identified, Cladosporium, is commonly found on both living and dead plant matter, and a xylanse from Cladosporium cladosporioides was isolated and characterized (Hong et al., 2011). A fourth genus, Wilcoxina is ectendomycorrhizal, penetrating the root cells of its host plant. The previously reported capacity to slowly degrade pectin and cellulose was proposed to facilitate root penetration (Redlak et al., 2001; Trevor et al., 2001), and a hemicellulolytic capacity may further contribute to this. This is the first report of hemicellulolytic activity in Ascomycota belonging to Venturia, Capronia, Xenopolyscytalum and Pseudeurotiaceae. A study of 14 Capronia spp. found that they were unable to grow on cellulose or lignin and concluded that the group was unable to function as primary degraders of plant biomass (Untereiner and Malloch, 1999); however, our results suggest that this may not be universally true of Capronia spp.
The hemicellulolytic bacterial populations identified appear to be similarly distributed between IDF and PP, whereas the fungal populations are not. This appears to be true for the other four ecozones as well. The bacterial populations account for a similar proportion of the overall bacterial communities in all six ecozones, about two percent (Figure 3), and the most abundant of these populations generally have similar abundances across the ecozones (Figure 4). Furthermore, the strains isolated on xylan, which correspond to two hemicellulolytic populations identified by SIP, were isolated from multiple ecozones (jack pine and PP). By contrast, the fungal populations identified are most abundant in the IDF and PP ecozones (Figures 3 and 5), which were the sources of soil for the microcosms. Thus, it appears that distinct hemicellulolytic fungal populations dominate each ecozone. This pattern is consistent with the previously reported patchy distribution of total fungi in IDF and sub-boreal spruce ecozones and soils from other mixed coniferous forests (Kranabetter and Wylie, 1998; Tedersoo et al., 2003; Hartmann et al., 2012). Based on these observations, it might be possible to infer distributions of hemicellulolytic bacterial populations in all six ecozones on the basis of the in situ pyrotag libraries, whereas it is possible to do so for hemicellulolytic fungal populations in only the IDF and PP ecozones.
A greater proportion of the organic versus mineral layer community, both bacterial and fungal populations, is hemicellulolytic in the IDF and PP ecozones. This difference is indicated by in situ pyrotag data (Figure 3) and the steeper slope for the organic versus mineral layer in the correlation of hemicellulose assimilation and total biomass (Figure 1). The pyrotag data indicate that bacterial hemicellulytic populations are slightly, but significantly more abundant in the organic layer, whereas fungal populations are almost three-times more abundant in the organic layer. This difference is consistent with the expectation that lignocellulose decomposition occurs to a greater extent in the organic versus mineral layer. However, our SIP-PLFA experiments demonstrate that the communities in the mineral layer are clearly capable of metabolizing hemicellulose. This is consistent with hemicellulose degradation associated with root decomposition in the mineral layer. Our results from microcosms do not necessarily reflect in situ rates of hemicellulose degradation, rather they reflect the size and metabolic potential of hemicellulolytic populations in situ.
Timber harvesting significantly affected hemicellulolytic populations in the IDF and PP ecozones in a manner that persisted for 10 or more years. However, these effects were limited to subtle shifts in community composition. Relative abundances of only a few populations were clearly affected by harvesting. Most notably, the most abundant bacterial hemicellulolytic population (OTU13), affiliated with Burkholderia, exhibited a consistent decrease in relative abundance with harvesting (Figure 7). Harvesting did not alter the proportion of the overall community, that is, hemicellulolytic, nor did it alter the evenness of hemicellulolytic populations. The magnitude of change due to harvesting was much smaller than community differences between soil layers and between ecozones (Table 2). Variable effects of harvesting on soil characteristics and tree regeneration have been observed throughout the LTSP study (Kranabetter et al., 2006). Hemicellulolytic populations were more severely affected by harvesting in PP versus the IDF ecozone (Supplementary Figure S5). Interestingly, this is one of few ecozones in which tree regeneration was affected by harvesting method, being stimulated by increasing levels of OM removal (Fleming et al., 2006; Ponder et al., 2012). Of the six ecozones sampled for this study, the PP ecozone has an extreme precipitation regime, with the highest annual precipitation but the lowest precipitation during the warmest season (Table 1). Harvesting effects, in terms of variability explained by harvesting treatments, on hemicellulolytic soil populations in the IDF ecozone are of a similar magnitude as previously reported effects on the overall bacterial and fungal community composition in that ecozone (Hartmann et al., 2012). Thus, there is no indication that the hemicellulolytic populations were disproportionately affected by harvesting, as originally hypothesized.
Varying the level of OM removal during harvesting modulated the effect of harvesting on hemicellulolytic soil populations in the IDF and PP ecozones. This conclusion is supported by both canonical analysis of principal coordinates (Figure 6) and PERMANOVA (Table 2). Again, differences among the three harvested treatments amounted to subtle shifts in relative abundances. There were very few populations with consistent and significant responses to varied levels of OM removal, even when each soil layer of each ecozone was separately examined. In particular, differences were minimal between effects of stem-only harvesting (OM1) versus whole-tree harvesting (OM2). Stem-only harvesting is intended to leave some of the harvesting residue to maintain soil fertility and structure (Powers et al., 2005). However, for soil hemicellulolytic populations, those residues did little to offset the disturbance associated with harvesting. Accordingly, previous studies have shown only small differences in the effects of OM1 versus OM2 on overall bacterial and fungal communities (Hartmann et al., 2012) and small or no differences in effects on soil chemistry and tree growth (Ponder et al., 2012). The OM3 treatment, involving removal of whole trees plus forest floor, is a very severe disturbance, which was previously found to impact the overall soil microbial community due to the loss of nutrients and habitat (Simard et al., 2003; Hartmann et al., 2012). This treatment eliminated all above-ground woody biomass and was expected to have an extreme effect on hemicellulolytic populations. Of course, the hemicellulolytic populations in the organic layer were removed by the OM3 treatment, but the effect on those in the mineral layer was comparable to the change associated with OM1 and OM2. In those ecozones where the organic layer had regenerated sufficiently in the OM3 treatment to permit sampling (PP and loblolly pine), hemicellulolytic populations were affected by harvesting to a larger extent than those in OM1 and OM2 treatments (Supplementary Table S2).
Varying the level of OM removal during harvesting did not detectably alter potential rates of hemicellulose degradation and assimilation (Supplementary Figure S4). Thus, the observed differences in hemicellulolytic population structure among harvested treatments did not appear to change the hemicellulolytic potential of the community. In accordance with this, most LTSP Study sites so far lack detectable effects of harvesting treatments on tree regeneration or soil properties (Conlin and Driessche, 2000; Powers et al., 2005, 2012). Timber-harvesting effects on hemicellulose degradation have never been directly investigated, but litter decomposition has previously been reported to be unaffected by harvesting in the IDF and sub-boreal spruce ecozones (Prescott et al., 2000). However, tree regeneration at these sites is in an early phase, and as the trees grow, changing factors will limit that growth. It would be informative to monitor the soil communities over the long term to determine whether the relatively subtle effects of harvesting on the soil community ultimately correspond to effects on forest regeneration. It would be also be informative to determine whether the soil communities in harvested treatments eventually converge with those of the corresponding references.
Overall, this study identified major hemicellulolytic bacterial and fungal populations in forest soils, and showed that the bacterial populations, but not the fungal ones, are similarly distributed across multiple ecozones. Forest harvesting caused long-term changes to the structure of hemicellulolytic populations in the IDF and PP ecozones, but it did not appear to substantially alter the total biomass or metabolic potential of hemicellulolytic microorganisms. It remains to be determined whether changes in community structure due to harvesting eventually have consequences for ecosystem processes that affect forest productivity or resilience to disturbance.
Acknowledgments
This study was supported by grants from Genome Canada and Genome BC. Kendra R Maas was supported by a postdoctoral fellowship from the Tula foundation and Roland C Wilhelm was supported by the Alexander Graham Bell Canada Graduate Scholarship (CGS) from the Natural Sciences and Engineering Research Council of Canada. We thank participants of the LTSP Study, including S Berch, C Bulmer, MD Busse, WK Chapman, RL Fleming, PW Hazlett, G Hope, JM Kranabetter, DM Morris, DA Scott, K Webster, for access to field sites, assistance sampling and soil data. We thank Larisa McNeil for soil processing and DNA extractions. We thank Pedro Dimitriu and Erick Cardenas for helpful discussions during data analysis. We also thank the contribution of scientists at the McGill University and Genome Quebec Innovation Center, Montreal, Canada for their 454 pyrosequencing service.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Biely P, Krátký Z, Kocková-Kratochvílová A, Bauer Š. (1978). Xylan-degrading activity in yeasts: growth on xylose, xylan and hemicelluloses. Folia Microbiol 23: 366–371. [DOI] [PubMed] [Google Scholar]
- Bligh E, Dyer WJ. (1959). A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Canadell JG, Raupach MR. (2008). Managing forests for climate change mitigation. Science 320: 1456–1457. [DOI] [PubMed] [Google Scholar]
- Chávez R, Bull P, Eyzaguirre J. (2006). The xylanolytic enzyme system from the genus Penicillium. J Biotechnol 123: 413–433. [DOI] [PubMed] [Google Scholar]
- Cheng C-L, Chang J-S. (2011). Hydrolysis of lignocellulosic feedstock by novel cellulases originating from Pseudomonas sp. CL3 for fermentative hydrogen production. Bioresour Technol 102: 8628–8634. [DOI] [PubMed] [Google Scholar]
- Coenye T, Vandamme P. (2003). Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol 5: 719–729. [DOI] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlin T, Driessche Rvd. (2000). Response of soil CO2 and O2 concentrations to forest soil compaction at the long-term soil productivity sites in central British Columbia. Can J Soil Sci 80: 625–632. [Google Scholar]
- de Castro VHL, Schroeder LF, Quirino BF, Kruger RH, Barreto CC. (2013). Acidobacteria from oligotrophic soil from the Cerrado can grow in a wide range of carbon source concentrations. Can J Microbiol 59: 746–753. [DOI] [PubMed] [Google Scholar]
- Dixon P. (2003). VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- Doran JW. (2002). Soil health and global sustainability: translating science into practice. Agric Ecosyst Environ 88: 119–127. [Google Scholar]
- Estrada-de los Santos P, Vinuesa P, Martínez-Aguilar L, Hirsch AM, Caballero-Mellado J. (2013). Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr Microbiol 67: 51–60. [DOI] [PubMed] [Google Scholar]
- FAO. (2010). Rome: Global Forest Resources Assessment. [Google Scholar]
- Fleming RL, Powers RF, Foster NW, Kranabetter JM, Scott DA, Ponder F Jr et al. (2006). Effects of organic matter removal, soil compaction, and vegetation control on 5-year seedling performance: a regional comparison of Long-Term Soil Productivity sites. Can J Forest Res 36: 529–550. [Google Scholar]
- Frostegård Å, Tunlid A, Bååth E. (1991). Microbial biomass measured as total lipid phosphate in soils of different organic content. J Microbiol Methods 14: 151–163. [Google Scholar]
- Ghio S, Di Lorenzo GS, Lia V, Talia P, Cataldi A, Grasso D et al. (2012). Isolation of Paenibacillus sp. and Variovorax sp. strains from decaying woods and characterization of their potential for cellulose deconstruction. Int J Biochem Mol Biol 3: 352. [PMC free article] [PubMed] [Google Scholar]
- Harazono K, Yamashita N, Shinzato N, Watanabe Y, Fukatsu T, Kurane R. (2003). Isolation and characterization of aromatics-degrading microorganisms from the gut of the lower termite Coptotermes formosanus. Biosci Biotechnol Biochem 67: 889–892. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Howes CG, VanInsberghe D, Yu H, Bachar D, Christen R et al. (2012). Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J 6: 2199–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-Y, Kim Y-H, Jung M-H, Jo C-W, Choi J-E. (2011). Characterization of Xylanase of Cladosporium cladosporioides H1 isolated from Janggyeong Panjeon in Haeinsa Temple. Mycobiology 39: 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DW, Curtis PS. (2001). Effects of forest management on soil C and N storage: meta analysis. Forest Ecol Manag 140: 227–238. [Google Scholar]
- Keenan RJ, Kimmins JP. (1993). The ecological effects of clear-cutting. Environ Rev 1: 121–144. [Google Scholar]
- Khan H, Chung EJ, Jeon CO, Chung YR. (2013. a). Mucilaginibacter gynuensis sp. nov., isolated from rotten-wood. Int J Syst Evol Microbiol 63: 3225–3231. [DOI] [PubMed] [Google Scholar]
- Khan H, Chung EJ, Kang DY, Jeon CO, Chung YR. (2013. b). Mucilaginibacter jinjuensis sp. nov., with xylan-degrading activity. Int J Syst Evol Microbiol 63: 1267–1272. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Kranabetter JM, Wylie T. (1998). Ectomycorrhizal community structure across forest openings on naturally regenerated western hemlock seedlings. Can J Botany 76: 189–196. [Google Scholar]
- Kranabetter JM, Sanborn P, Chapman BK, Dube S. (2006). The contrasting response to soil disturbance between lodgepole pine and hybrid white spruce in subboreal forests. Soil Sci Soc Am J 70: 1591–1599. [Google Scholar]
- Kurz WA, Stinson G, Rampley GJ, Dymond CC, Neilson ET. (2008). Risk of natural disturbances makes future contribution of Canada's forests to the global carbon cycle highly uncertain. Proc Natl Acad Sci 105: 1551–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki ML, Idrees A, Leung KT, Qin W. (2012). Newly isolated and characterized bacteria with great application potential for decomposition of lignocellulosic biomass. J Mol Microbiol Biotechnol 22: 156–166. [DOI] [PubMed] [Google Scholar]
- Mohana S, Shah A, Divecha J, Madamwar D. (2008). Xylanase production by Burkholderia sp. DMAX strain under solid state fermentation using distillery spent wash. Bioresour Technol 99: 7553–7564. [DOI] [PubMed] [Google Scholar]
- Moore-Kucera J, Dick R. (2008). PLFA profiling of microbial community structure and seasonal shifts in soils of a douglas-fir chronosequenprofiling of microbial community structure and seasonal shifts in soils of a douglas-fir chronosequence. Microb Ecol 55: 500–511. [DOI] [PubMed] [Google Scholar]
- Morosoli R, Zalce E, Durand S. (1993). Secretion of a Cryptococcus albidus xylanase in Pichia stipitis resulting in a xylan fermenting transformant. Curr Genet 24: 94–99. [DOI] [PubMed] [Google Scholar]
- Nakatsu CH, Hristova K, Hanada S, Meng X-Y, Hanson JR, Scow KM et al. (2006). Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol 56: 983–989. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW et al. (2007). DNA stable-isotope probing. Nat Protoc 2: 860–866. [DOI] [PubMed] [Google Scholar]
- Oravecz O, Elhottová D, Krištůfek V, Šustr V, Frouz J, Tříska J et al. (2004). Application of ARDRA and PLFA analysis in characterizing the bacterial communities of the food, gut and excrement of saprophagous larvae ofPenthetria holosericea (Diptera: Bibionidae): a pilot study. Folia Microbiol 49: 83–93. [DOI] [PubMed] [Google Scholar]
- Pankratov TA, Dedysh SN. (2010). Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int J Syst Evol Microbiol 60: 2951–2959. [DOI] [PubMed] [Google Scholar]
- Pérez J, Muñoz-Dorado J, de la Rubia T, Martínez J. (2002). Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol 5: 53–63. [DOI] [PubMed] [Google Scholar]
- Ponder F Jr, Fleming RL, Berch S, Busse MD, Elioff JD, Hazlett PW et al. (2012). Effects of organic matter removal, soil compaction and vegetation control on 10th year biomass and foliar nutrition: LTSP continent-wide comparisons. Forest Ecol Manag 278: 35–54. [Google Scholar]
- Powers R. (1999). On the sustainable productivity of planted forests. New Forests 17: 263–306. [Google Scholar]
- Powers RF, Andrew Scott D, Sanchez FG, Voldseth RA, Page-Dumroese D, Elioff JD et al. (2005). The North American long-term soil productivity experiment: Findings from the first decade of research. Forest Ecol Manag 220: 31–50. [Google Scholar]
- Powers RF. (2006). Long-Term Soil Productivity: genesis of the concept and principles behind the program. Can J Forest Res 36: 519–528. [Google Scholar]
- Prescott CE, Blevins LL, Staley CL. (2000). Effects of clear-cutting on decomposition rates of litter and forest floor in forests of British Columbia. Can J Forest Res 30: 1751–1757. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat SR, Männistö MK, Starovoytov V, Goodwin L, Nolan M, Hauser L et al. (2014). Complete genome sequence of Granulicella tundricola type strain MP5ACTX9(T), an Acidobacteria from tundra soil. Stand Genom Sci 9: 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlak K, Dahm H, Ciesielska A, Strzelczyk E. (2001). Enzymatic activity of ectendomycorrhizal fungi. Biol Fert Soils 33: 83–90. [Google Scholar]
- Salles JF, Van Veen JA, Van Elsas JD. (2004). Multivariate analyses of Burkholderia species in soil: effect of crop and land use history. Appl Environ Microbiol 70: 4012–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez FG, Scott DA, Ludovici KH. (2006). Negligible effects of severe organic matter removal and soil compaction on loblolly pine growth over 10 years. Forest Ecol Manag 227: 145–154. [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16 S rRNA-Based Studies. PLoS ONE 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL. (2011). Assessing and improving methods used in operational taxonomic unit-based approaches for 16 S rRNA gene sequence analysis. Appl Environ Microbiol 77: 3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shallom D, Shoham Y. (2003). Microbial hemicellulases. Curr Opin Microbiol 6: 219–228. [DOI] [PubMed] [Google Scholar]
- Simard SW, Jones MD, Durall DM, Hope GD, Stathers RJ, Sorensen NS et al. (2003). Chemical and mechanical site preparation: effects on Pinus contorta growth, physiology, and microsite quality on grassy, steep forest sites in British Columbia. Can J Forest Res 33: 1495–1515. [Google Scholar]
- Sinsabaugh RL, Antibus RK, Linkins AE, McClaugherty CA, Rayburn L, Repert D et al. (1993). Wood decomposition: nitrogen and phosphorus dynamics in relation to extracellular enzyme activity. Ecology 74: 1586–1593. [Google Scholar]
- Song J, Cho J-C. (2007). Methylibium aquaticum sp. nov., a betaproteobacterium isolated from a eutrophic freshwater pond. Int J Syst Evol Microbiol 57: 2125–2128. [DOI] [PubMed] [Google Scholar]
- Štursová M, Žifčáková L, Leigh MB, Burgess R, Baldrian P. (2012). Cellulose utilization in forest litter and soil: identification of bacterial and fungal decomposers. FEMS Microbiol Ecol 80: 735–746. [DOI] [PubMed] [Google Scholar]
- Swift MJ, Heal OW, Anderson JM. (1979) Decomposition in Terrestrial Ecosystems vol. 5 University of California Press: Los Angeles, CA, USA. [Google Scholar]
- Talia P, Sede SM, Campos E, Rorig M, Principi D, Tosto D et al. (2012). Biodiversity characterization of cellulolytic bacteria present on native Chaco soil by comparison of ribosomal RNA genes. Res Microbiol 163: 221–232. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Kõljalg U, Hallenberg N, Larsson K-H. (2003). Fine scale distribution of ectomycorrhizal fungi and roots across substrate layers including coarse woody debris in a mixed forest. New Phytol 159: 153–165. [DOI] [PubMed] [Google Scholar]
- Trevor EY, Egger KN, Peterson LR. (2001). Ectendomycorrhizal associations–characteristics and functions. Mycorrhiza 11: 167–177. [Google Scholar]
- Turner S, Pryer KM, Miao VP, Palmer JD. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukar Microbiol 46: 327–338. [DOI] [PubMed] [Google Scholar]
- Untereiner WA, Malloch D. (1999). Patterns of substrate utilization in species of capronia and allied black yeasts: ecological and taxonomic implications. Mycologia 91: 417–427. [Google Scholar]
- Van Der Heijden MGA, Bardgett RD, Van Straalen NM. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11: 296–310. [DOI] [PubMed] [Google Scholar]
- VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam SJ, Mohn WW. (2015). Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J e-pub ahead of print 24 April 2015; doi:10.1038/ismej.2015.54. [DOI] [PMC free article] [PubMed]
- Varnaitċ R, Raudonienċ V. (2008). Destruction of hemicellulose in rye straw by micromycetes. Ekologija 54: 169–172. [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M et al. (2009). Threegenomes from the phylum acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75: 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. (1991). 16 S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc 18: 315–322. [Google Scholar]
- Wilhelm R, Szeitz A, Klassen TL, Mohn WW. (2014). Sensitive, efficient quantitation of 13c-enriched nucleic acids via ultrahigh-performance liquid chromatography–tandem mass spectrometry for applications in stable isotope probing. Appl Environ Microbiol 80: 7206–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Zheng Y, Yu X, Yu L, Gao D, Chen S. (2013). Lignocellulosic biomass as a carbohydrate source for lipid production by Mortierella isabellina. Bioresour Technol 128: 385–391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.