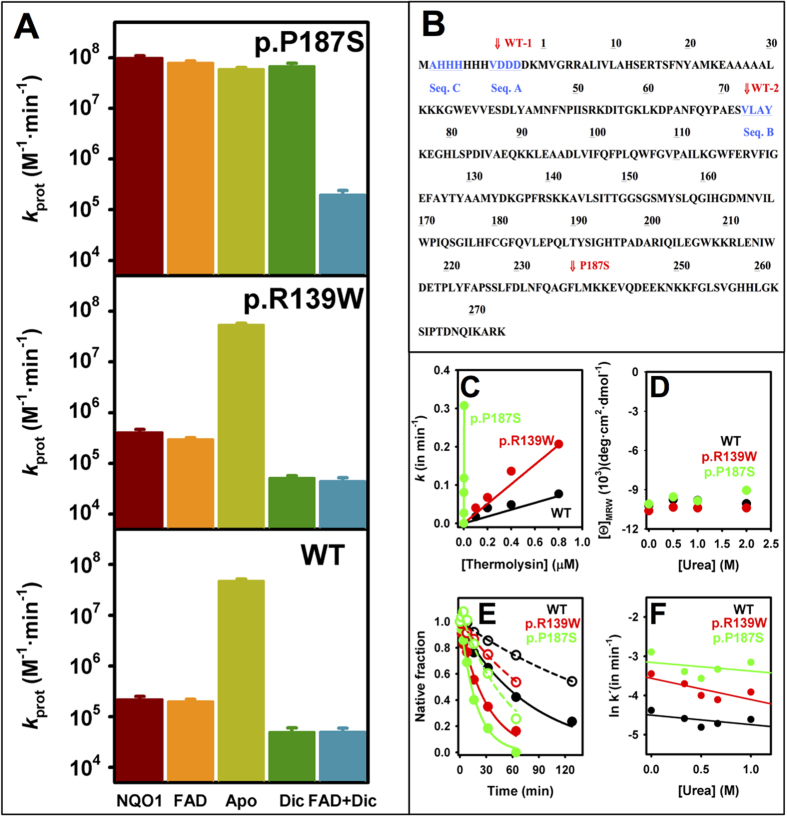
Figure 3. Proteolysis probes the local dynamics at N-terminal and C-terminal domains of NQO1 and the effects of polymorphisms and ligand binding.
(A) Proteolysis rate constants of different NQO1 enzymes under different experimental conditions: NQO1 (as purified), FAD (in FAD excess), Apo (FAD withdrawn), Dic (in dicoumarol excess) and FAD + Dic (in FAD and dicoumarol excess). (B) Cleavage sites mapped onto the sequence of the WT NQO1 construct used in our study. Numbering corresponding to the NQO1 sequence excluding the N-terminal tag. Seqs (A–C) are N-terminal sequences determined for selected proteolysis products (highlighted in blue and underlined). The primary cleavage sites are labeled in red. See the main text for additional details. (C–F) proteolysis probes the local dynamics of NQO1 proteins. (C) The linear response of proteolysis rate constants with protease concentration shows that the proteolysis step is rate-limiting under these conditions; (D) Far-UV CD signals at 220 nm after 4 h incubation at selected urea concentrations (protein concentration 6 μM in monomer); (E) Proteolysis of NQO1 enzymes as holo-proteins after incubation with 1 M urea for at least two hours (open symbols) or without urea (closed symbols). Thermolysin concentrations were 0.1 μM (WT and p.R139W) and 1 nM (p.P187S). (F) Dependence of the proteolysis rate constants at different urea concentrations after correction by the urea effect on thermolysin activity reported by28 (k´). The equilibrium m values are −0.14 ± 0.12 (WT), −0.30 ± 0.15 (p.R139W) and −0.12 ± 0.21 (p.P187S), in kcal·mol−1·M−1.