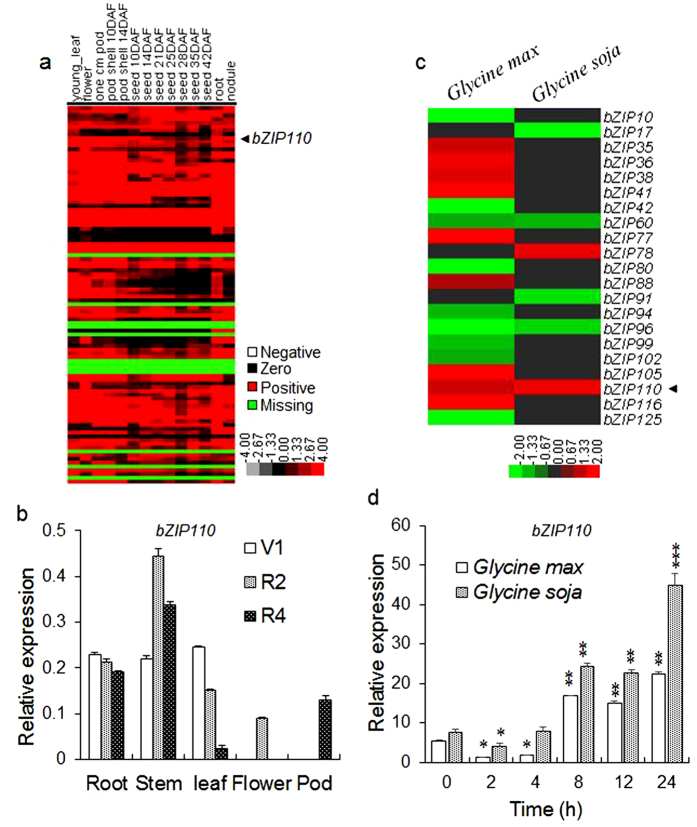
Figure 2. Transcript abundance profiles of GmbZIP genes.
(a) In silico transcript abundance data for full length GmbZIP genes in young leaves, flowers, one cm pods, pod shells at 10 days after flowering (DAF), pod shells at 14 DAF, seeds at 10 DAF, seeds at 14 DAF, seeds at 21 DAF, seeds at 25 DAF, seeds at 28 DAF, seeds at 35 DAF, seeds at 42 DAF, roots and nodules was downloaded from SOYBASE. The legend (−4–4) indicates gene expression level. (b) Transcript abundance of GmbZIP110 in the roots, stems, leaves, flowers and pods of soybean at the first trifoliate (V1), full bloom (R2) and full pod (R4) stages. The experiments were carried out using qPCR. Different individual lowercase letters indicate significant differences as determined by one-way ANOVA followed by a Bonferroni post-hoc test. (c) Normalized transcript abundance (log2 ratio) of differentially expressed GmbZIP genes in a salt-tolerant genotype of Glycine soja (STGoGS) and a salt-sensitive genotype of Glycine max (SSGoGM) under 0 and 200 mM NaCl stress expressed as digital gene expression profiling (DGEP) data. The legend (−2–2) indicates the log2 ratio, which is the gene expression level under 200 mM NaCl treatment relative to that under 0 mM NaCl treatment. (d) Differential transcript abundance of GmbZIP110 in the roots of STGoGS and SSGoGM under 0 and 200 mM NaCl stress. P-values were calculated using Student’s t-test. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with 0 h.