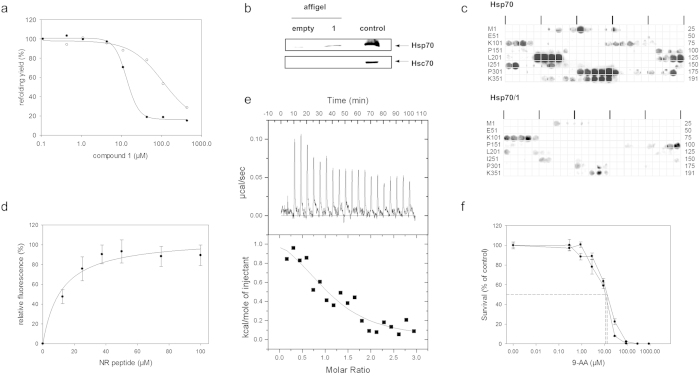
Figure 1. Characterization of the Hsp70 inhibition by 1.
(a) Inhibitory effect of 1 on the reactivation of GdmCl-denatured firefly luciferase by Hsp70. Refolding was initiated by dilution of 2.08 μM unfolded luciferase into refolding buffer containing Hsp70 (closed circles) or Hsc70 (open circles). The drawn line was calculated using an IC50 of 45 μM for Hsp70 and 363 μM for Hsc70. Each data point represents the average of duplicate determinations which differed not more than 10%. (b) Derivative 1 interacted with Hsp70 but not with Hsc70 in HeLa cell lysate. Affigel-10 beads pre-incubated with 1 were incubated with HeLa cell lysate. Bound proteins were analyzed by SDS-PAGE and immunoblotting using anti-Hsp70 and anti-Hsc70 antibodies, respectively. As controls, a loading control and affigel-10 beads not pre-incubated with 1 were used. (c) Binding of Hsp70 to p53 derived peptides was inhibited by 1. Binding was detected by incubating a cellulose-bound peptide array of p53-derived 12mer peptides with Hsp70 alone (upper panel) and inhibition of binding was observed in the additional presence of 1 (lower panel). (d) The peptide NRLLLTG competed with compound 1 for binding to Hsp70. Analysis was performed in triplicate by a fluorescence-based competition experiment. The continuous lines represent the best fits of the data to a single binding site model. Error bars represent SD. (e) Isothermal titration of Hsp70 with 1. Upper panel: raw titration data; lower panel: integrated enthalpies. A solution of 20 μM Hsp70 (initial concentration) was titrated at 20 °C with 200 μM 1and fitted to a single-site binding model, resulting in a KD of 5.6 ± 3.1 μM, with stoichiometry N = 1.1, ΔHITC = 1.3 ± 0.3 kcal mol−1, and TΔSITC = 8.3 kcal mol−1. (f) The effect of acridizinium derivatives on cell survival using the clonogenic assay. EC50 were determined to be 11.2 μM for 1 and 13.1 μM for 3.