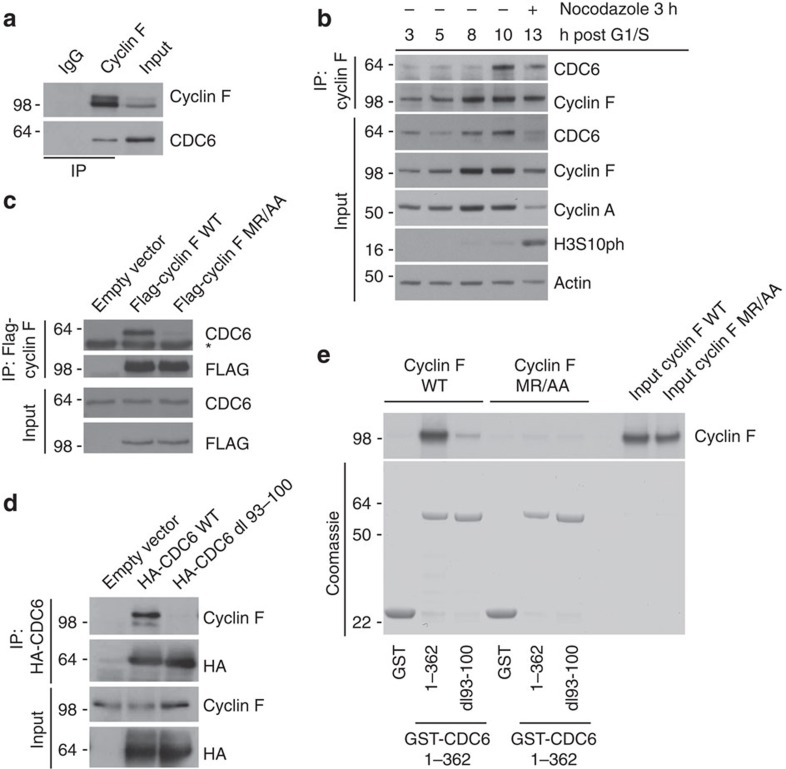
Figure 1. CDC6 and Cyclin F interact through defined molecular motifs.
(a) Whole-cell extracts of U2OS cells were generated, lysates were immunoprecipitated (IP) with anti-Cyclin F antibody and IPs were analysed by immunoblotting as indicated. (b) U2OS cells were synchronized at G1/S by using a double-thymidine block before release into fresh medium. Whole-cell extracts were generated at the indicated time points, lysates were IP with anti-Cyclin F antibody and IPs were analysed by immunoblotting as indicated. (c) HEK-293T cells were transfected with an empty vector, FLAG-tagged wild-type Cyclin F (WT) or FLAG-tagged mutant Cyclin F(MR/AA). Whole-cell extracts were IP with anti-FLAG resin, and IPs were analysed by immunoblotting as indicated. *Notes a cross reacting band in the CDC6 immunoblot. (d) HEK-293T cells were transfected with an empty vector, HA-tagged wild-type CDC6 (WT) or HA-tagged mutant CDC6 (dl 93–100). Whole-cell extracts were IP with anti-HA resin, and IPs were analysed by immunoblotting as indicated. (e) The interaction between recombinant WT and dl 93–100 mutant GST-CDC6 fragments (amino acids 1–362) and in-vitro translated and S35-methionine-labelled WT and MR/AA mutant Cyclin F was tested. GST alone was used as a negative control. The GST-fusion proteins were precipitated by glutathione-agarose affinity chromatography and the interaction with the in vitro translated S35-labelled proteins was visualized by autoradiography.