Figure 3.
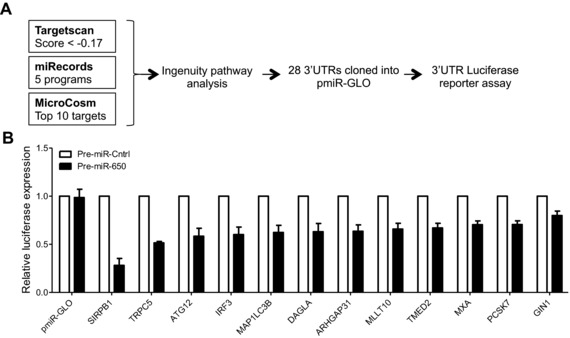
In silico prediction of miR‐650 targets and screening by luciferase reporter assay. (A) Schematic representation of the workflow used to select candidate miR‐650 target genes. A list of candidate targets genes was compiled by considering the top third of all predictions performed by TargetScan (total context score ≤ −0.17) 20, results of the miRecords metasearch as well as the top‐scoring miR‐650 targets retrieved from MicroCosm. Full‐length 3′UTR sequences of 28 putative target genes were cloned into pmiR‐GLO reporter vector and tested for their response to miR‐650 by dual luciferase reporter assay. (B) All pmiR‐GLO constructs significantly (p < 0.05) regulated following cotransfection with miR‐650 pre‐miR in dual luciferase screen. pmiR‐GLO empty vector served as negative control. Firefly activity was normalized to Renilla and is expressed relative to control pre‐miRNA. Measurements were performed in triplicates or quadruplicates and data are shown as mean ± SEM and are pooled from three independent experiments. Statistical significance was tested using a paired Student's t‐test. All shown targets were significant, p < 0.05.
