C. elegans SORF-1 and SORF-2 and their mammalian homologs WDR91 and WDR81 maintain appropriate PtdIns3P levels in early-to-late endosome conversion by forming a complex with the Beclin1 subunit of the PI3K complex.
Abstract
Phosphatidylinositol 3-phosphate (PtdIns3P) plays a central role in endosome fusion, recycling, sorting, and early-to-late endosome conversion, but the mechanisms that determine how the correct endosomal PtdIns3P level is achieved remain largely elusive. Here we identify two new factors, SORF-1 and SORF-2, as essential PtdIns3P regulators in Caenorhabditis elegans. Loss of sorf-1 or sorf-2 leads to greatly elevated endosomal PtdIns3P, which drives excessive fusion of early endosomes. sorf-1 and sorf-2 function coordinately with Rab switching genes to inhibit synthesis of PtdIns3P, allowing its turnover for endosome conversion. SORF-1 and SORF-2 act in a complex with BEC-1/Beclin1, and their loss causes elevated activity of the phosphatidylinositol 3-kinase (PI3K) complex. In mammalian cells, inactivation of WDR91 and WDR81, the homologs of SORF-1 and SORF-2, induces Beclin1-dependent enlargement of PtdIns3P-enriched endosomes and defective degradation of epidermal growth factor receptor. WDR91 and WDR81 interact with Beclin1 and inhibit PI3K complex activity. These findings reveal a conserved mechanism that controls appropriate PtdIns3P levels in early-to-late endosome conversion.
Introduction
Delivery of intracellular cargoes to lysosomes involves maturation of early endosomes through homotypic fusion, early-to-late endosome conversion, and heterotypic fusion of late endosomes with lysosomes. These processes are mainly controlled by endosome-specific Rab GTPases and phosphatidylinositides (Stenmark, 2009; Huotari and Helenius, 2011). The Rab GTPases Rab5 and Rab7 are required for fusion of early and late endosomes, respectively. Rab5 recruitment to early endosomes is facilitated by its guanine nucleotide exchange factor Rabex-5, which converts Rab5 into an active GTP-bound form that interacts with Rabaptin-5 (Horiuchi et al., 1997). Rabaptin-5 further enhances the guanine nucleotide exchange factor activity of Rabex-5, establishing a positive feedback loop of Rab5 activation, leading to rapid recruitment of other Rab5 effectors including Vps34, a class III phosphatidylinositol 3-kinase (PI3K), and class C core vacuole/endosome tethering (CORVET)/homotypic fusion and vacuole protein sorting (HOPS), a tethering complex (Christoforidis et al., 1999b; Lippé et al., 2001; Murray et al., 2002; Peplowska et al., 2007; Plemel et al., 2011). This leads to assembly of trans-SNARE complexes between membranes to promote fusion and maturation of early endosomes (Wickner, 2010; Balderhaar et al., 2013). Early-to-late endosome conversion requires replacement of Rab5 with Rab7, which is controlled by a complex consisting of Mon1/SAND-1 and Ccz1/CCZ-1 (Rink et al., 2005; Nordmann et al., 2010; Poteryaev et al., 2010). By recognizing the PtdIns3P level and size of early endosomes, SAND-1 displaces RABX-5/Rabex-5 from the endosome membrane, which probably interrupts the positive feedback loop of Rab5 activation. The SAND-1–CCZ-1 complex also recruits and activates Rab7 on endosome membranes (Poteryaev et al., 2007, 2010). Once activated, GTP-bound Rab7 (GTP-Rab7) recruits effectors including TBC-2, a Rab5 GTPase activating protein (GAP), which terminates Rab5 activity (Li et al., 2009; Chotard et al., 2010), and the HOPS complex, which mediates fusion of late endosomes.
The characteristic phosphatidylinositide of early endosomes, phosphatidylinositol 3-phosphate (PtdIns3P), plays key roles in the endosome-lysosome pathway (Di Paolo and De Camilli, 2006; Schink et al., 2013). PtdIns3P promotes homo- or heterotypic fusion of early endosomes through PtdIns3P-binding proteins, such as EEA1, Rabenosyn-5, and Phafin2 (Christoforidis et al., 1999a; Nielsen et al., 2000; Gengyo-Ando et al., 2007; Subramanian et al., 2010; Pedersen et al., 2012). PtdIns3P is also important for cargo sorting to lysosomes or recycling back to the trans-Golgi network (Futter et al., 2001; Henne et al., 2011; Seaman, 2012). PtdIns3P is mainly generated on endosomes by the class III PI3K complex, which consists of Vps34, p150/Vps15, and Beclin 1/Atg6 and is recruited by GTP-Rab5 (Christoforidis et al., 1999b; Murray et al., 2002; Funderburk et al., 2010; Huotari and Helenius, 2011). PtdIns3P is most abundant on early endosomes and endosomal carrier vesicles, intermediates between early and late endosomes. PtdIns3P is not obviously present on multivesicular late endosomes (Gillooly et al., 2000), probably because of dephosphorylation by myotubularin family phosphatases or conversion by PIKfyve/Fab1 into the late endosome–specific phosphatidylinositide, PtdIns(3,5)P2, or degradation in the endosomal lumen (Wurmser and Emr, 1998; Huotari and Helenius, 2011; Schink et al., 2013). Thus, control of PtdIns3P generation and elimination is crucial for early-to-late endosome conversion and the subsequent switch of endosome identities and endosome-to-lysosome trafficking. The interactions of Rab5 or Rab7 with PI3K or myotubularin phosphatases are thought to be critical for PtdIns3P turnover (Christoforidis et al., 1999b; Murray et al., 2002; Stein et al., 2003; Cao et al., 2007, 2008); however, other factors or mechanisms regulating endosomal PtdIns3P levels remain to be identified.
Caenorhabditis elegans is an excellent model organism for studying membrane trafficking (Sato et al., 2014). C. elegans has six macrophage-like cells (coelomocytes) that actively undergo fluid-phase endocytosis and contain endosomes and lysosomes that are easily distinguished with differential interference contrast (DIC) optics or organelle-specific fluorescent markers. Importantly, molecules controlling the fusion and fission of endosomes and lysosomes as well as early-to-late endosome conversion are well conserved (Fares and Greenwald, 2001a,b; Grant and Sato, 2006; Sato et al., 2014). Previously, we found that deletion of the CORVET/HOPS subunit VPS-18 caused severe defects in endosome/lysosome fusion in C. elegans coelomocytes (Xiao et al., 2009). To search for additional components of the endosome-lysosome pathway, we performed vps-18 suppressor screens and identified two novel factors, SORF-1 and SORF-2, which are essential for appropriate maintenance of PtdIns3P levels in early-to-late endosome conversion. SORF-1 and SORF-2 form a complex that further interacts with the BEC-1 subunit of the PI3K complex. Deletion of sorf-1 or sorf-2 led to enrichment of BEC-1 on early endosomes and enhanced PI3K complex activity, resulting in prolonged existence of endosomal PtdIns3P and delay in early-to-late endosome conversion. WDR91 and WDR81, the mammalian homologs of SORF-1 and SORF-2, also act in complex with Beclin1 to regulate endosomal PtdIns3P levels by suppressing PI3K activity. These findings reveal a conserved negative regulatory mechanism by which endosomal PtdIns3P is controlled in early-to-late endosome conversion.
Results
Mutations in sorf-1 and sorf-2 partially suppress defective endosome fusion in CORVET/HOPS mutants
To identify additional regulators in the endosome-lysosome pathway, we performed genetic screens for genes whose loss of function could partially rescue the endosome/lysosome fusion defect in coelomocytes in C. elegans vps-18(tm1125) CORVET/HOPS mutants (Xiao et al., 2009). ZK563.5 and F52C9.1 were identified from an RNAi screen of genes on chromosomes V and X and an EMS mutagenesis screen, respectively (Fig. S1, A–C). Compared with vps-18(tm1125) single mutants, double mutants of vps-18 with a deletion of ZK563.5, tm3855, have larger endosomes and lysosomes, as indicated by the endosome marker RME-8::RFP (Zhang et al., 2001) and the lysosome marker LMP-1::GFP (Fig. 1, A–C). Double mutants of vps-18(tm1125) with the yq4 mutant of F52C9.1 also have larger endosomes and lysosomes than vps-18 single mutants (Figs. 1, A–C; and S1 C). This was confirmed with a deletion mutant of F52C9.1, tm5210 (Fig. S1, B and C). Deletion of ZK563.5 or RNAi of F52C9.1 also suppressed defective endosome fusion in deletion mutants of other HOPS subunits except vps-16 and -33.1 (Fig. S1 E).
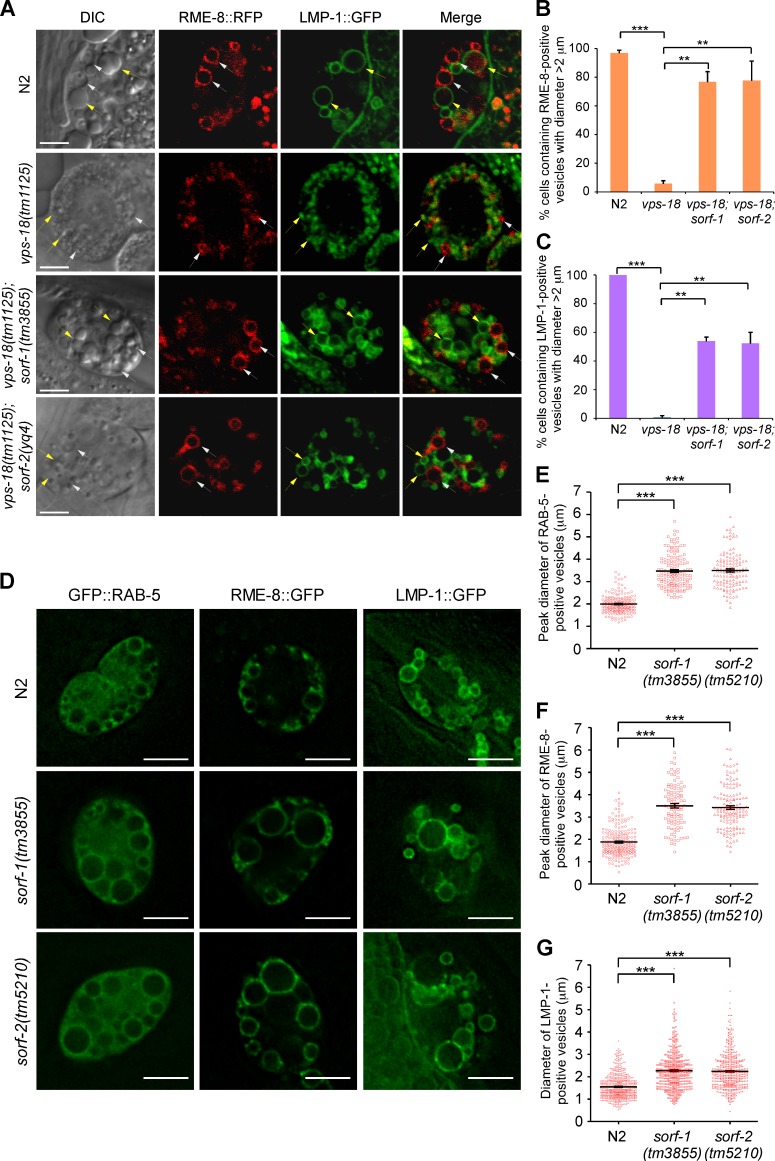
Figure 1.
Loss of sorf-1 and sorf-2 partially suppresses defective organelle fusion in vps-18 mutants and causes enlargement of endosomes. (A) Representative confocal images of endosomes and lysosomes in coelomocytes in N2, vps-18(tm1125), vps-18(tm1125);sorf-1(tm3855), and vps-18(tm1125);sorf-2(yq4) animals. DIC images are shown together with RME-8::RFP (white arrows), LMP-1::GFP (yellow arrows), and merged fluorescence images. Bars, 5 µm. (B and C) Proportion of cells containing enlarged RME-8::RFP- or LMP-1::GFP-positive vesicles in animals with the indicated genotype. ≥70 coelomocytes (one coelomocyte/animal) were scored. (D) Representative images of endosomes labeled with GFP-RAB-5 or RME-8::GFP and lysosomes labeled with LMP-1::GFP in coelomocytes of the indicated animals. Bars, 5 µm. (E and F) Peak sizes of endosomes positive for GFP::RAB-5 (n > 100) and RME-8::GFP (n > 100). (G) Sizes of LMP-1::GFP-positive lysosomes (n > 500). For all quantifications, error bars represent SEM derived from three independent experiments. **, P < 0.01; ***, P < 0.001. In E–G, each symbol represents an endosome or lysosome; the horizontal line indicates the mean peak diameter.
To examine the endosome/lysosome phenotypes of deletion mutants of these two genes, we monitored endosomes and recorded their diameters when they reached their largest sizes (peak diameters). In ZK563.5(tm3855) and F52C9.1(tm5210) mutant coelomocytes, early endosomes labeled by GFP-tagged RAB-5 (GFP::RAB-5) or RME-8 (RME-8::GFP) were significantly larger than in wild type (N2; Fig. 1, D–F). Late endosomes/lysosomes, labeled with the lysosome membrane protein LMP-1::GFP, were also larger in the mutants (Fig. 1, D and G). Collectively, these findings suggest that both genes play critical roles in the endosome-lysosome pathway. ZK563.5 and F52C9.1 were named sorf-1 (suppressor of organelle fusion 1) and sorf-2, respectively. sorf-1 encodes a protein with similarity to the N-terminus of the mammalian WD40 repeat–containing protein WDR91 (Fig. S1 D). The protein encoded by sorf-2 contains a Beach (Beige and CHS) domain and a WD40-repeat region and shows homology to the mammalian WD40 repeat–containing protein WDR81 (Fig. S1 D). Mutation of WDR81 is associated with cerebellar ataxia, mental retardation, and disequilibrium syndrome-2 (Gulsuner et al., 2011). However, the functions of WDR81 and WDR91 are unknown.
Loss of sorf-1 and sorf-2 causes endosome enlargement and elevated endosomal PtdIns3P levels
To study the role of sorf-1 and sorf-2 in the endosome-lysosome pathway, we examined PtdIns3P levels on endosomes using GFP-tagged 2xFYVE (2xFYVE::GFP) or EEA-1 (GFP::EEA-1), both of which specifically bind PtdIns3P without affecting endosome size (Fig. S1 F; Gillooly et al., 2000; Balklava et al., 2007). 2xFYVE::GFP signals were significantly higher on enlarged endosomes in sorf-1(tm3855) and sorf-2(tm5210) mutants than in wild type (Fig. 2, A and B). GFP::EEA-1 was also enriched on endosomes in sorf-1 or sorf-2 mutants (Fig. 2, A and C). These results suggest that loss of sorf-1 or sorf-2 results in significantly increased endosomal PtdIns3P levels.
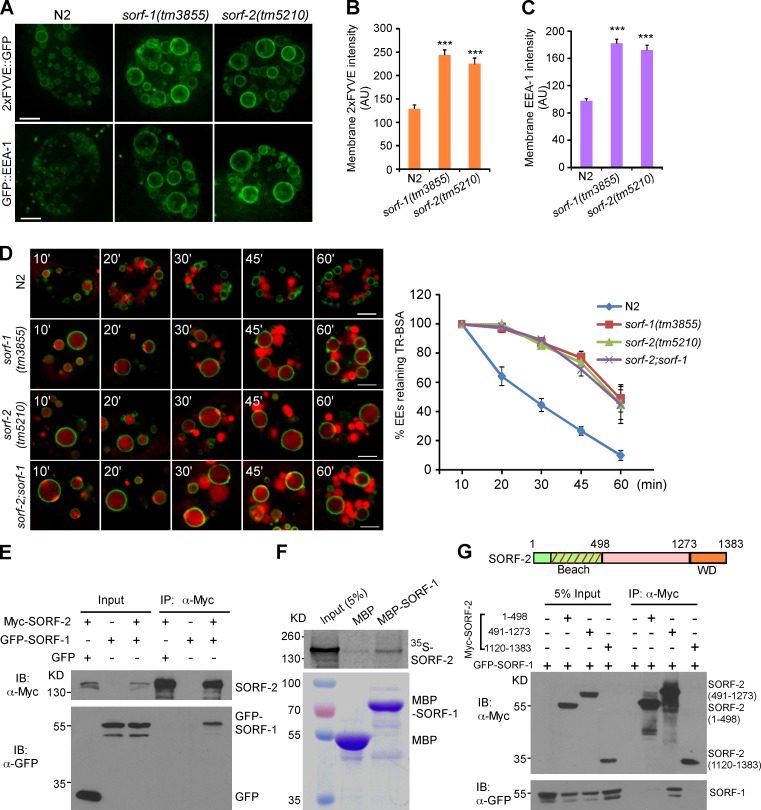
Figure 2.
Loss of sorf-1 and sorf-2 leads to PtdIns3P enrichment and defective endosomal transport. (A) Representative projection images of 2xFYVE::GFP or GFP::EEA-1 on early endosomes in N2, sorf-1(tm3855), and sorf-2(tm5210) coelomocytes. Images were obtained with the same exposure time. Projections were made for the whole cell. (B and C) Intensity of 2xFYVE::GFP or GFP::EEA-1 (arbitrary units [AU]) on endosomes. ≥300 endosomes were measured per genotype. N2 and mutants were compared with t tests. Error bars represent SEM. ***, P < 0.001. (D) Time course of endosomal TR-BSA trafficking in coelomocytes in animals of the indicated genotypes. TR-BSA was injected into the body cavity and examined at the indicated time points after injection (left). Bars, 5 µm. Early endosomes (EEs) were quantified for retention of TR-BSA (right). 60 or more endosomes were scored at each time point for each genotype. Error bars represent SEM, and data are from three independent experiments. (E–G) Interaction of SORF-1 and SORF-2. Myc-tagged SORF-2 (E) or its different subregions (G) were coexpressed with GFP-SORF-1 in HEK293 cells. IPs were performed with Myc antibody, and precipitated proteins were detected with Myc and GFP antibodies. 35S-labeled SORF-2 was incubated with MBP-SORF-1 and precipitated with maltose-binding beads (F).
We then investigated whether the enlarged, PtdIns3P-enriched endosomes affected endosome cargo transport by injecting Texas red–BSA (TR-BSA) into the C. elegans body cavity and monitoring its entry into and exit from 2xFYVE::GFP-positive early endosomes (Zhang et al., 2001). In wild-type coelomocytes, TR-BSA entered into early endosomes 10 min after injection but exited early endosomes quickly, and was seen in very few early endosomes 60 min after injection (Fig. 2 D). In sorf-1(tm3855) and sorf-2(tm5210) mutants, early endosomes accumulated TR-BSA 10 min after injection; however, far more endosomes retained TR-BSA at all time points observed after injection (Fig. 2 D). Thus, cargo transport from early to late endosomes was significantly delayed in the mutants.
We next asked whether sorf-1 and sorf-2 function in the same genetic pathway. Endosomal PtdIns3P levels, as measured by 2xFYVE::GFP signals, were not enhanced in sorf-2;sorf-1 double mutants compared with their single mutants (Fig. 2, A–C; and see Fig. 3 E). TR-BSA transport in double-mutant coelomocytes was also similar to that in single mutants (Fig. 2 D). In addition, GFP-SORF-1, but not GFP, was coimmunoprecipitated with Myc-SORF-2 when coexpressed in HEK293 cells (Fig. 2 E). Likewise, maltose-binding protein (MBP)-fused SORF-1 pulled down 35S-labeled SORF-2 in vitro, suggesting a direct interaction between SORF-1 and SORF-2 (Fig. 2 F). Further coimmunoprecipitations (coIPs) revealed that GFP-SORF-1 interacted only with the central region (aa 491–1273) of SORF-2 (Fig. 2 G). These data suggest that SORF-1 and SORF-2 act together in the endosome-lysosome pathway.
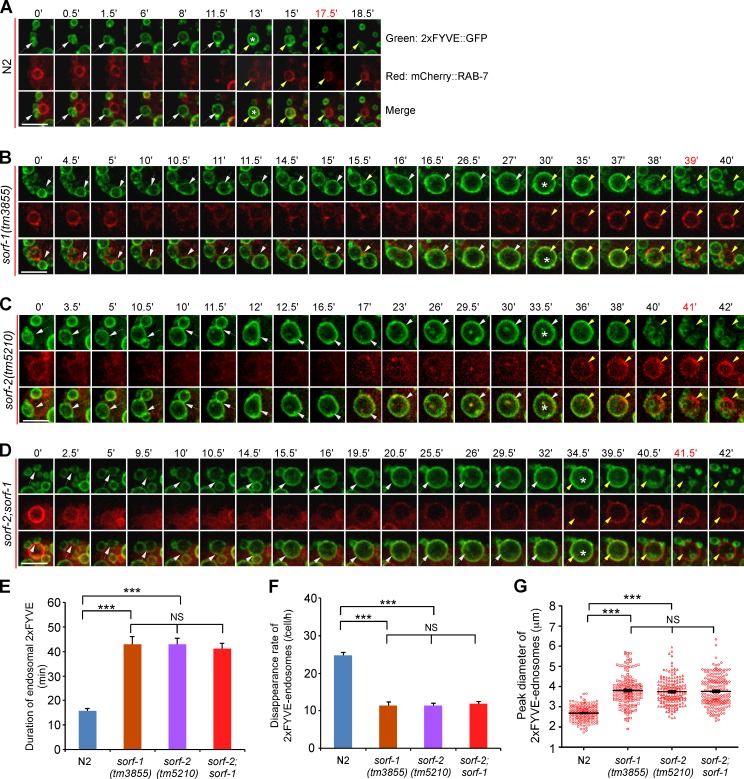
Figure 3.
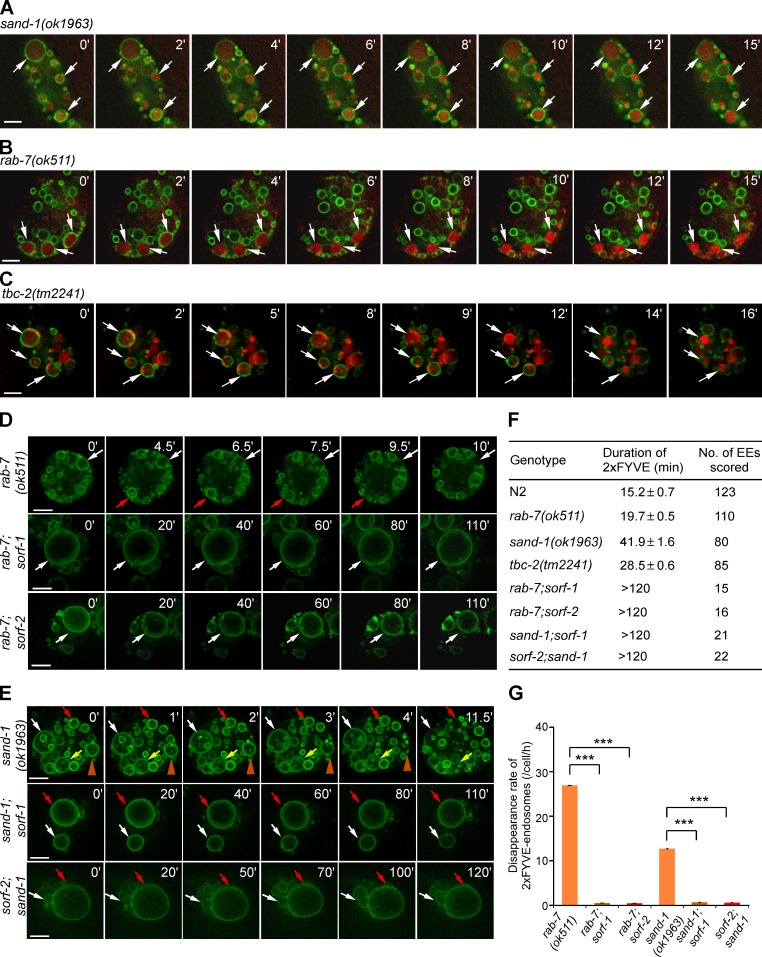
Loss of sorf-1 and sorf-2 delays early-to-late endosome conversion. (A–D) Time course of dynamic changes of PtdIns3P (2xFYVE::GFP) and RAB-7 (mCherry::RAB-7) on endosomes in N2 (A), sorf-1(tm3855) (B), sorf-2(tm5210) (C), and sorf-2(tm5210);sorf-1(tm3855) (D) coelomocytes. *, Time points at which endosomes reach peak sizes. White and yellow arrows indicate the chased endosomes before and after enrichment of RAB-7, respectively. Bars, 5 µm. (E) Duration of 2xFYVE::GFP on endosomes. ≥18 endosomes were examined for each genotype. (F) Rate of disappearance of 2xFYVE::GFP-positive endosomes. ≥12 coelomocytes were examined. (G) Peak sizes of endosomes (n > 100) in A–D. Each symbol represents an endosome that was followed. ***, P < 0.001; NS, no statistical significance.
Endosome conversion is delayed in sorf-1 and sorf-2 mutants
We next investigated the fate of the enlarged early endosomes in sorf-1 and sorf-2 deletion mutants by simultaneously monitoring the endosomal dynamics of PtdIns3P, which is labeled by 2xFYVE::GFP, and the appearance of RAB-7, which is tagged with mCherry (mCherry::RAB-7). In wild type, the PtdIns3P signal and the size of early endosomes increased from 0 to 13 min; the PtdIns3P signal was then stable for a short time (13–15 min) before disappearing quickly (Fig. 3 A and Video 1). RAB-7 progressively appeared on early endosomes, reaching a peak as the PtdIns3P signal began to decrease (Fig. 3 A, 13–17.5 min). Thus, conversion of early to late endosomes is marked by a strong decrease or turnover of endosomal PtdIns3P and appearance of RAB-7. In sorf-1(tm3855) and sorf-2(tm5210) single or double mutants, early endosomes kept fusing to form very big vesicles until RAB-7 was recruited and 2xFYVE::GFP signals disappeared (Fig. 3, B–D; and Video 1). The mean duration of endosomal PtdIns3P in single or double mutants of sorf-1 and sorf-2 was more than two times longer than wild type, whereas the disappearance rate of 2xFYVE::GFP-positive endosomes (total number of endosomes undergoing conversion/cell/h) was more than two times lower (Fig. 3, E and F). The mean peak size of early endosomes before disappearance of PtdIns3P was significantly larger in sorf-1 and sorf-2 single or double mutants (Fig. 3 G). These data further indicate that loss of sorf-1 or sorf-2 prolongs the existence of endosomal PtdIns3P, resulting in a delay in early-to-late endosome conversion and consequently defective endosome cargo transport. To consolidate this conclusion, we monitored the dynamic change of RAB-5 (GFP::RAB-5) and RAB-7 (mCherry::RAB-7) on endosomes. In N2 coelomocytes, the progressive disappearance of RAB-5 and enrichment of RAB-7 on endosomes normally occurred within ∼15 min (Fig. S2, A and D; and Video 2). In contrast, it took >30 min for RAB-5 to disappear from and RAB-7 to become enriched on endosomes in sorf-1(tm3855) or sorf-2(tm5210) coelomocytes (Fig. S2, B–D; and Video 2). These results further confirm that loss of sorf-1 or sorf-2 leads to a significant delay in early-to-late endosome conversion.
sorf-1 and sorf-2 act in parallel with genes required for endosomal Rab switching
To further pinpoint the role of SORF-1 and SORF-2 in regulating PtdIns3P on endosomes, we constructed double mutants of sorf-1 or sorf-2 with genes required for Rab switching in early-to-late endosome conversion, including rab-7, sand-1, and ccz-1. Loss of function of these genes leads to a failure in endosome conversion, arresting endocytic vesicles at the early endosome stage (Poteryaev et al., 2007; Nieto et al., 2010). Surprisingly, 2xFYVE::GFP-labeled early endosomes were greatly enlarged in hypodermal cells in double mutants of rab-7(ok511) with sorf-1(tm3855) or sorf-2(tm5210) (Fig. 4 A), and each coelomocyte in the double mutants contained a gigantic 2xFYVE::GFP-positive vacuole (Fig. 4, B and E). RNAi depletion of rab-7 in sorf-1 and -2 deletion mutants similarly caused gigantic vacuoles, a phenotype that was strongly rescued by expression of Flag-tagged SORF-1 or Myc-tagged SORF-2 (Fig. S3, A and B). Abnormal giant vacuoles positive for 2xFYVE::GFP were also observed in coelomocytes in double mutants of sand-1(ok1963) with sorf-1(tm3855) or sorf-2(yq4) (Fig. 4, C and E). These data indicate that simultaneous inhibition of endosomal Rab switching with deletion of sorf-1 or sorf-2 caused more severe enlargement of early endosomes. We confirmed the identity of these early endosomes by showing that they were negative for the lysosomal enzyme ASP-1 fused with dsRed (ASP-1::dsRED; Nakae et al., 2010; Fig. S3 C) and for LysoTracker red (Fig. S3 D). However, they were positive for the lysosome channel CUP-5 fused with mCherry (mCherry::CUP-5; Fig. 4, B and C) and LMP-1::GFP (Fig. S3 C), which might be because of trapping of these membrane proteins on the enlarged endosomes.
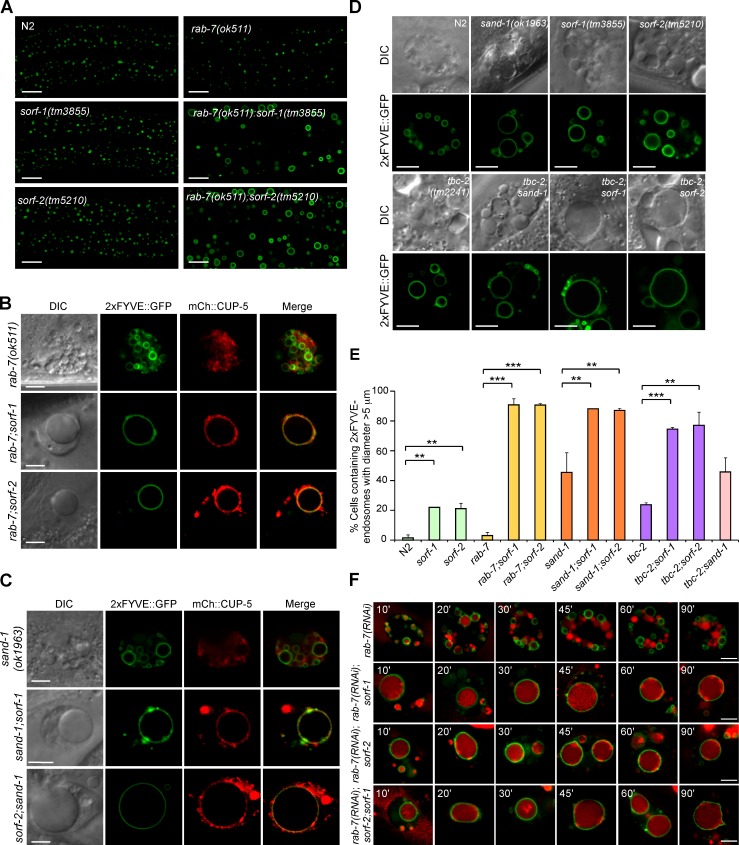
Figure 4.
Inhibiting the endosomal Rab switch in sorf-1 or sorf-2 mutants leads to formation of gigantic early endosomes. (A) Early endosomes (2xFYVE::GFP-vesicles) are enlarged in hypodermal cells in double mutants of rab-7(ok511) with sorf-1(tm3855) or sorf-2(tm5210). (B and C) Early endosomes, labeled with 2xFYVE::GFP, are enlarged in coelomocytes of double mutants of rab-7(ok511) (B) or sand-1(ok1963) (C) with sorf-1(tm3855) and sorf-2(tm5210). The lysosomal protein CUP-5 is tagged with mCherry (mCh::CUP-5). (D) Early endosomes are enlarged in coelomocytes of double mutants of tbc-2(tm2241) with sorf-1(tm3855) or sorf-2(tm5210) but not with sand-1(ok1963). (E) Proportion of cells with enlarged 2xFYVE::GFP-positive endosomes (diameter >5 µm). ≥70 coelomocytes from ≥70 animals (one cell/animal) were scored for each genotype. Data are from three independent experiments. **, P < 0.01; ***, P < 0.001. (F) Time course of endosomal trafficking of TR-BSA in coelomocytes of rab-7 RNAi–treated animals. Bars, 5 µm.
TBC-2 is a GAP of RAB-5, and its loss of function locks RAB-5 in an active GTP-bound form (GTP-RAB-5), which can act through Vps34 to promote PtdIns3P synthesis on endosomes or phagosomes (Li et al., 2009; Chotard et al., 2010). Double mutants of tbc-2 with sorf-1 or sorf-2 contain 2xFYVE::GFP-positive abnormal giant endosomes, similar to double mutants of rab-7 or sand-1 with sorf-1 or sorf-2 (Fig. 4, D and E). In contrast, sand-1;tbc-2 double mutants did not show additive enlargement of early endosomes (Fig. 4, D and E), suggesting that these genes act in the same genetic pathway (Poteryaev et al., 2007; Chotard et al., 2010). These findings suggest that sorf-1 and sorf-2 act in parallel to genes required for endosomal Rab switching. Consistent with this, TR-BSA persisted in the gigantic early endosomes in rab-7 RNAi-treated single or double mutants of sorf-1 and sorf-2 but exited normally from early endosomes in wild-type or rab-7(RNAi) animals (Figs. 4 F and 2 D).
Endosomal PtdIns3P turnover is abrogated in the absence of both SORF-1/SORF-2 function and Rab switching
We analyzed the duration of endosomal PtdIns3P in single mutants of genes required for Rab switching and their double mutants with sorf-1 or sorf-2 by time-lapse recording. Surprisingly, we found that endosomal 2xFYVE::GFP disappeared similarly in rab-7(ok511), sand-1(ok1963), and tbc-2(tm2241) deletion mutants when early endosomes grew to a certain size, although some early endosomes grew much larger and had prolonged existence of 2xFYVE::GFP in sand-1(ok1963) and tbc-2(tm2241) single mutants (Fig. 5, A–C and F; Poteryaev et al., 2007; Li et al., 2009; Chotard et al., 2010). These observations suggest that loss of endosomal Rab switching genes is not sufficient to prevent PtdIns3P turnover on endosomes. In contrast, in double mutants of sorf-1 or sorf-2 with rab-7 or sand-1, 2xFYVE::GFP persisted on the giant endosomes during the entire monitoring period (>2 h), and disappearance of 2xFYVE-positive endosomes was seldom observed (Fig. 5, D–G; and Videos 3 and 4). These data further suggest that SORF-1 and SORF-2 act in parallel with Rab switching genes (sand-1, ccz-1, rab-7, and tbc-2) to negatively regulate PtdIns3P on endosomes.
Figure 5.
Endosomal PtdIns3P persists in sorf-1 or sorf-2 mutants when Rab switch genes are deleted. (A–C) Dynamics of PtdIns3P (marked with 2xFYVE::GFP) in sand-1(ok1963) (A), rab-7(ok511) (B), and tbc-2(tm2241) (C) mutants. Arrows indicate disappearance of 2xFYVE::GFP from endosomes after injection of TR-BSA. (D and E) Time-lapse imaging of endosomal 2xFYVE::GFP in coelomocytes in rab-7(ok511) mutants and double mutants of rab-7 with sorf-1 or sorf-2 (D) and in sand-1(ok1963) mutants and double mutants of sand-1 with sorf-1 or sorf-2 (E). (F) Duration of endosomal 2xFYVE::GFP (mean ± SEM) in the indicated genotypes. EEs, early endosomes. (G) Disappearance rate of 2xFYVE::GFP-positive vesicles. ≥16 coelomocytes were scored per genotype. ***, P < 0.001.
PtdIns3P persistence leads to continuous fusion of endosomes
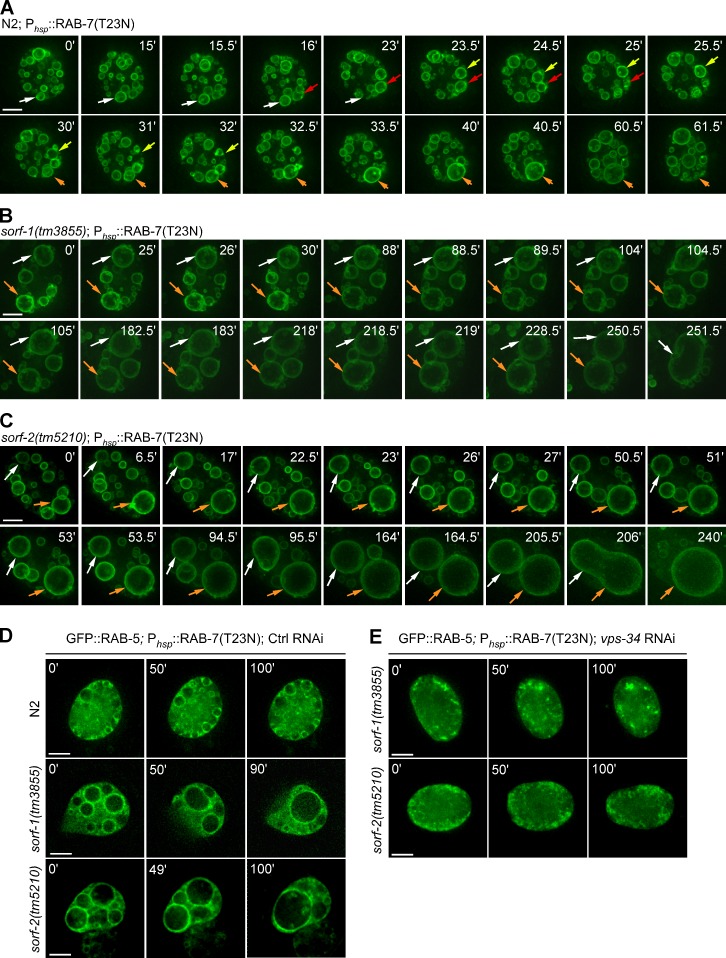
We next used a heat shock promoter to express a dominant-negative form of RAB-7, PhspRAB-7(T23N), in wild type and sorf-1 or sorf-2 mutants. After heat shock, we followed 2xFYVE::GFP-positive endosomes by time-lapse analysis. In wild type, early endosomes positive for 2xFYVE::GFP fused after RAB-7(T23N) was expressed, but no further fusion occurred once endosomes reached a certain size (Fig. 6 A, compare 0' with 61.5' after heat shock; and Video 5). In sorf-1(tm3855) and sorf-2(tm5210) animals, however, heat shock–induced RAB-7(T23N) expression caused continuous fusion of 2xFYVE::GFP-positive endosomes into a giant endosome (Fig. 6, B and C; and Video 5). No fission of early endosomes was observed during the monitoring period. Heat shock–induced RAB-7(T23N) similarly led to continuous fusion of endosomes labeled with GFP::RAB-5 in sorf-1 and sorf-2 mutants (Fig. 6 D), which was nevertheless abrogated by RNAi depletion of vps-34 (Fig. 6 E). These findings indicate that inhibition of both the endosomal Rab switching machinery and SORF-1 or SORF-2 function resulted in persistence of PtdIns3P, which in turn led to continuous fusion of early endosomes to form abnormally large organelles.
Figure 6.
PI3K-dependent persistence of PtdIns3P leads to continuous fusion of early endosomes. (A–C) Time-lapse analysis of RAB-7(T23N)-induced fusion of 2xFYVE::GFP-labeled early endosomes in N2 (A), sorf-1(tm3855) (B), and sorf-2(tm5210) (C) coelomocytes. Colored arrows indicate different endosomes that were followed. (D) Time-lapse images of RAB-7(T23N)-induced fusion of early endosomes labeled with GFP::RAB-5. (E) Time-lapse images of RAB-7(T23N)-induced fusion of early endosomes labeled with GFP::RAB-5 in vps-34 RNAi–treated sorf-1 and sorf-2 mutants. Bars, 5 µm.
SORF-1 and SORF-2 function through BEC-1 to regulate endosomal PtdIns3P
Our findings suggest that SORF-1 and SORF-2 are required for endosomal turnover of PtdIns3P in the absence of Rab switch genes. To understand this function, we examined the genetic interaction of sorf-1 or sorf-2 with phosphatases or kinases that promote or down-regulate endosomal PtdIns3P (Fig. S4 A). Deletion of sorf-1 or RNAi of rab-7 in mutants of genes encoding myotubularin family phosphatases (including mtm-1, -3, -5, -6, and -9; Fig. S4 B) or PtdIns3P 5-kinase (ppk-3; Fig. S4 C) did not show additive enlargement of endosomes. Deletion of the PtdIns(3,5)P2 phosphatase gene C34B7.2 or the class II PI3K gene piki-1 also had no effect on the endosome phenotype in sorf-1 or sorf-2 mutants treated with control RNAi or rab-7 RNAi (Fig. S4 D). These results suggest that SORF-1 and SORF-2 do not act through these phosphatases or kinases.
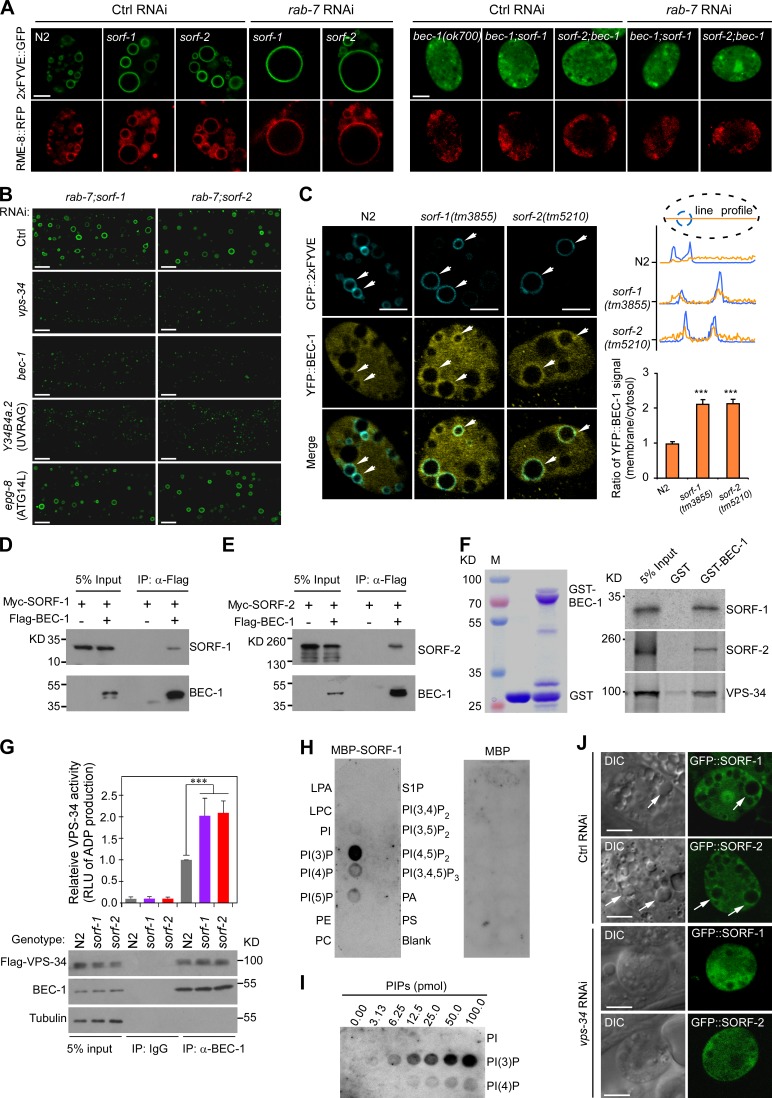
We next examined the requirement for the class III PI3K complex, which is composed of VPS-34, BEC-1, and VPS-15/p150. In bec-1(ok700) deletion mutants or its double mutants with sorf-1 or sorf-2, 2xFYVE::GFP became more evenly distributed in the cytoplasm, and RME-8::RFP-labeled early endosomes were much smaller than in wild type (Fig. 7 A). RNAi of rab-7 did not induce giant early endosomes in these double mutants (Fig. 7 A). These results suggest that BEC-1 is required for endosome enlargement and PtdIns3P persistence resulting from inhibition of both Rab switching and sorf-1 or sorf-2 functions. This is consistent with studies that the Beclin1/Atg6 subunit of the PI3K complex is required for endocytic trafficking in C. elegans and mammals (Thoresen et al., 2010; Ruck et al., 2011; McKnight et al., 2014). In double mutants of rab-7 with sorf-1 or sorf-2, RNAi depletion of vps-34, bec-1, or Y34B4a.2, which encodes the C. elegans homolog of UVRAG, strongly suppressed the enlargement of early endosomes in hypodermal cells (Fig. 7 B). In contrast, RNAi of epg-8, which encodes the C. elegans homolog of ATG14L, a subunit of the autophagy-specific PI3K complex (Lu et al., 2011), did not have a similar effect (Fig. 7 B). These data indicate that the endosome-specific PI3K complex is responsible for early endosome enlargement in single mutants of sorf-1 and sorf-2 as well as their double mutants with genes responsible for Rab switching.
Figure 7.
SORF-1 and SORF-2 act through BEC-1 to regulate endosomal PtdIns3P levels. (A) Images of endosomes colabeled with 2xFYVE::GFP and RME-8::RFP in coelomocytes of N2, sorf-1(tm3855), and sorf-2(tm5210) animals treated with control (Ctrl) RNAi or rab-7 RNAi (left), and in Ctrl RNAi- or rab-7 RNAi-treated bec-1(ok700) single mutants and double mutants of bec-1(ok700) with sorf-1(tm3855) or sorf-2(tm5210) (right). (B) RNAi of vps-34, bec-1, and Y34B4a.2 suppresses enlargement of early endosomes in hypodermal cells in double mutants of rab-7(ok511) with sorf-1(tm3855) or sorf-2(tm5210). (C) Left: images of BEC-1::YFP on early endosomes labeled with CFP::2xFYVE. Arrows indicate BEC-1::YFP enrichment. Top right: endosome membrane-to-cytoplasm ratio of BEC-1::YFP intensity. Dotted lines represent the coelomocyte boundary (black) and a CFP::2xFYVE-labeled endosome (blue). Fluorescence intensities of BEC-1::YFP (yellow curve) and CFP::2xFYVE (blue curve) were measured along a line across the endosome. Bottom right: mean membrane-to-cytoplasm ratio obtained from 35 line profiles in 20 coelomocytes for each genotype. ***, P < 0.001. (D and E) Flag-BEC-1 was coexpressed with Myc-SORF-1 (D) or Myc-SORF-2 (E) in HEK293 cells and immunoprecipitated with Flag antibody. Precipitated proteins were detected with Flag and Myc antibodies. (F) Purified GST or GST-BEC-1 (left) was incubated with 35S-labeled SORF-1, SORF-2, or VPS-34 and pulled down with glutathione-Sepharose beads. (G) Loss of sorf-1 or sorf-2 enhances PI3K complex activity. BEC-1 was immunoprecipitated from total lysates of N2, sorf-1(tm3855), and sorf-2(tm5210) animals expressing Flag-VPS-34. Precipitated proteins were detected with indicated antibodies (bottom). Equal amounts of precipitated proteins from each genotype were examined for PI3K activity by measuring relative light unit of luminescence (RLU) of ADP converted from ATP. Data representing mean ± SEM are from three independent experiments and are normalized to the PI3K activity of N2 animals (top). ***, P < 0.001. (H) MBP-SORF-1 (left) or MBP (right) was incubated with membrane strips dotted with indicated phospholipids. Bound protein was detected with MBP antibody. (I) Binding of MBP-SORF-1 to increasing amounts of PtdIns3P on strips. (J) DIC and fluorescence images of GFP::SORF-1 and GFP::SORF-2 in coelomocytes treated with Ctrl RNAi or vps-34 RNAi. Arrows indicate endosomes enriched in SORF-1 or SORF-2. Bars, 5 µm.
We next studied the PI3K complex in sorf-1 and sorf-2 mutant coelomocytes. YFP-fused BEC-1 (YFP::BEC-1) was distributed evenly in the cytoplasm in wild-type coelomocytes but was significantly enriched on, or in close proximity to, early endosomes in both sorf-1 and sorf-2 mutant coelomocytes (Fig. 7 C). The membrane-to-cytoplasm ratio of YFP::BEC-1 intensity was higher in sorf-1 and sorf-2 mutants than in wild type (Fig. 7 C), suggesting that SORF-1 and SORF-2 may prevent BEC-1 from accessing early endosomes. When coexpressed in HEK293 cells, Flag-BEC-1 and Myc-tagged SORF-1 or SORF-2 were coimmunoprecipitated by one another (Figs. 7, D and E; and S5, A and B). In pull-down assays, GST-fused BEC-1 pulled down 35S-labeled SORF-1, SORF-2, and VPS-34 (Fig. 7 F). No interactions were found between VPS-34 and SORF-1 or SORF-2 in coIP assays (Fig. S5, C and D). In addition, neither SORF-1 nor SORF-2 interacted with RAB-5, RAB-7, or their GTP- or GDP-bound forms in coIP assays (Fig. S5, E–H), excluding the possibility that SORF-1 or SORF-2 functions directly through RAB-5 or RAB-7. Thus, SORF-1 and SORF-2 likely inhibit PtdIns3P levels by specifically interacting with BEC-1. To test this, we immunoprecipitated BEC-1 from lysates of N2, sorf-1(tm3855), and sorf-2(tm5210) animals expressing Flag-VPS-34 and measured PI3K activity. Loss of sorf-1 or sorf-2 did not affect the BEC-1/VPS-34 interaction, because similar levels of Flag-VPS-34 were coprecipitated with BEC-1 in N2, sorf-1(tm3855), and sorf-2(tm5210) animals (Fig. 7 G). However, the activities of BEC-1-VPS-34 complexes from sorf-1 and sorf-2 mutants are nearly two times that from N2 animals (Fig. 7 G). In addition, MBP-SORF-1, but not MBP, specifically binds to PtdIns3P in a dose-dependent manner in vitro (Fig. 7, H and I), and GFP-fused SORF-1 or SORF-2 are enriched on or in close proximity to endosomes, although they are also seen in the cytoplasm (Fig. 7 J). The endosomal enrichment of GFP::SORF-1/SORF-2 was abolished with vps-34 RNAi (Fig. 7 J). Consistent with this, we found that Flag-SORF-1 or Myc-SORF-2 expressed in C. elegans were present in the soluble fraction but not in the membrane fraction (Fig. S5 J). These data suggest that the SORF-1–SORF-2 complex likely binds to endosomal PtdIns3P and acts through BEC-1 to suppress PI3K complex activity, thereby negatively regulating endosomal PtdIns3P levels.
Human WDR91 and WDR81 are recruited to endosomes and are important for endosomal trafficking
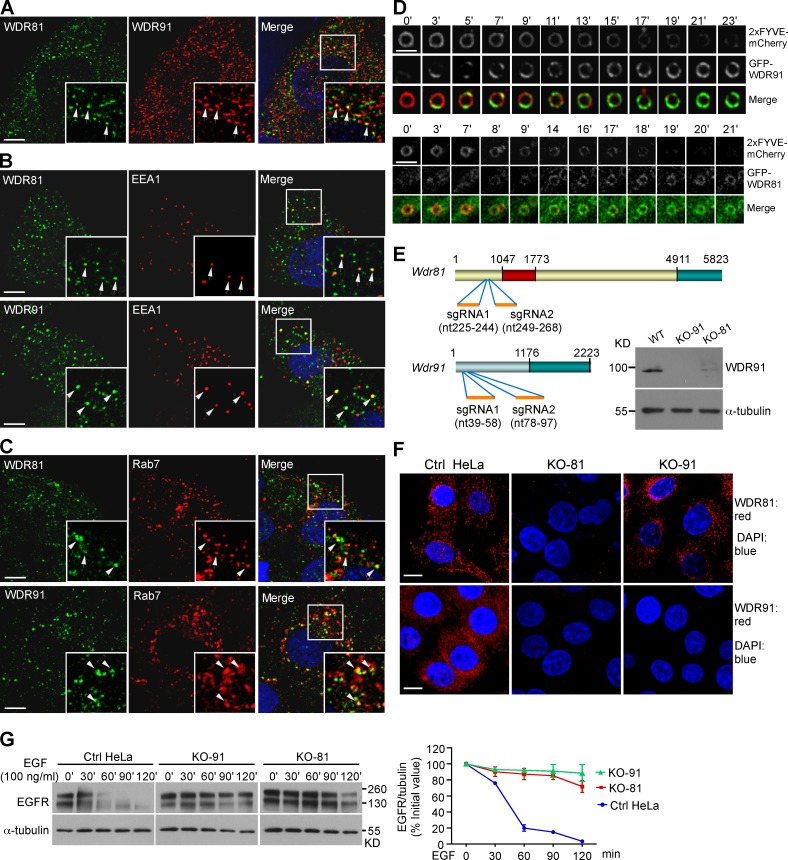
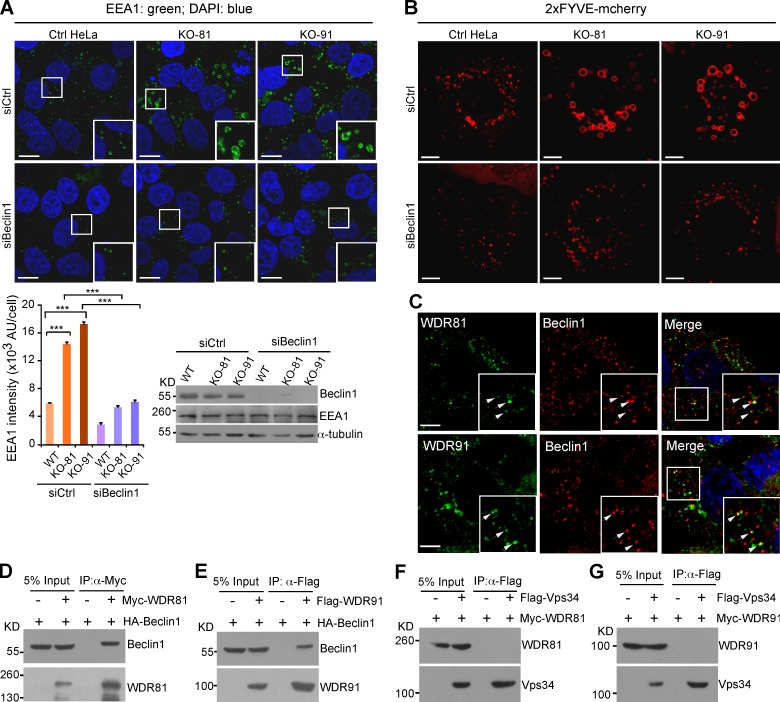
We investigated the function of WDR91 and WDR81, the mammalian homologs of SORF-1 and SORF-2. Endogenous WDR91 substantially colocalized with WDR81, and ectopically expressed Myc-WDR81 coimmunoprecipitated with Flag-WDR91 (Figs. 8 A and S5 I), suggesting that these proteins act in a complex. Next, we examined whether they localize to endosomes by immunostaining. Both endogenous WDR81 and WDR91 partially colocalized with the early endosome protein EEA1 (Fig. 8 B) or the late endosome protein Rab7 (Fig. 8 C). Live-cell imaging showed that endosomal PtdIns3P, indicated by 2xFYVE-mCherry, progressively disappeared, whereas WDR91 (GFP-WDR91) or WDR81 (GFP-WDR81) appeared on endosomes (Fig. 8 D; and Video 6). Neither protein appears to be stably associated with membranes (Fig. S5 J). To further test the role of WDR91 and WDR81 in endosomal trafficking, we generated WDR91 and WDR81 knockout (KO) cells using CRISPR/Cas9 (Fig. 8, E and F). The WDR91 protein level was strongly decreased in WDR81 KO cells, suggesting that WDR81 may be important for WDR91 stability (Fig. 8, E and F). Lysosome-mediated degradation of epidermal growth factor receptor (EGFR) was significantly impaired in WDR91 and WDR81 KO cells (Fig. 8 G), confirming a role for these two factors in endolysosomal trafficking.
Figure 8.
Human WDR91 and WDR81 are recruited to endosomes and are important for endolysosomal trafficking. (A) Colocalization of WDR91 with WDR81 (arrows) in immunostained HeLa cells. Insets show magnified (1.8×) views of the box in the merged image. (B and C) Coimmunostaining of WDR91 or WDR81 with EEA1 (B) or Rab7 (C). Insets show magnified (1.8×) views of the boxes in the merged images. Arrows indicate colocalization of WDR91 or WDR81 with EEA1 or Rab7. Bars: (A–C) 5 µm. (D) Time course of 2xFYVE::mCherry and GFP-WDR91 (top) and GFP-WDR81 (bottom) on endosomes. Bars, 2 µm. (E) Top: sgRNA targeting sites in WDR81 and WDR91 cDNAs. Numbers indicate nucleotides (nt) starting from the first ATG. Bottom right: Western blot of WDR91 in control (Ctrl), WDR91 knockout (KO-91), and WDR81 knockout (KO-81) HeLa cells. (F) Immunostaining of WDR91 or WDR81 in Ctrl, KO-91, and KO-81 cells. Red, WDR91 or WDR81 antibody; blue, DAPI. Bars, 10 µm. (G) Ctrl, KO-91, and KO-81 cells were treated with EGF for 10 min, and then EGFR levels were examined by Western blot (left). EGFR levels were quantified with ImageJ software (right). Data representing mean ± SEM are from three independent experiments.
WDR91 and WDR81 act in complex with Beclin1
To understand the role of WDR91 and WDR81 in endosomal dynamics, we examined early endosomes by immunostaining the early endosomal protein EEA1. Endogenous EEA1 was enriched on the enlarged endosomes in WDR81 or WDR91 KO cells, although total EEA1 levels remained the same as in control HeLa cells (Fig. 9 A). Given that EEA1 is a PtdIns3P-binding factor, this finding suggested that loss of WDR81 or WDR91 might result in elevated endosomal PtdIns3P. Consistent with this idea, an increase in endosome size was seen when endosomes were labeled with 2xFYVE-mCherry (Fig. 9 B), similar to our observations in C. elegans. This effect is probably not caused by an increase in macropinocytosis, because no obvious difference was detected between control HeLa cells and WDR91 or WDR81 KO cells in internalizing dextran 405 without or with EGF, which reflects constitutive or growth factor–induced macropinocytosis, respectively (Schnatwinkel et al., 2004; Kerr and Teasdale, 2009; Fig. S5, K–M). Importantly, both the endosomal enrichment of EEA1 and the enlargement of 2xFYVE-mCherry–positive endosomes seen in WDR91 or WDR81 KO cells were strongly suppressed by siRNA knockdown of Beclin1 (Fig. 9, A and B). Thus, loss of WDR91 or WDR81 caused a Beclin1-dependent increase in endosomal PtdIns3P and enlargement of early endosomes. Supporting this notion, we found that endogenous WDR81 and WDR91 both partially colocalize with Beclin1 (Fig. 9 C). Furthermore, HA-Beclin1, but not Vps34, was coimmunoprecipitated with Myc-WDR81 or Flag-WDR91 when coexpressed in HEK293 cells (Fig. 9, D–G).These results suggest that WDR91 and WDR81, like C. elegans SORF-1 and SORF-2, negatively regulate endosomal PtdIns3P by interacting with Beclin1.
Figure 9.
WDR91 and WDR81 act in complex with Beclin1 to affect endosomal PtdIns3P levels. (A) Immunostaining of endogenous EEA1 in control (Ctrl), KO-81, and KO-91 HeLa cells treated with control siRNA (siCtrl) and Beclin1 siRNA (siBeclin1; left). Boxed regions are magnified (2×) in insets. Bars, 15 µm. Bottom left: EEA1 intensity on endosomes (arbitrary units [AU]) in ≥100 cells. ***, P < 0.001. (bottom right) Western blot of Beclin1 and EEA1 in cells treated with siBeclin1 or siCtrl. (B) Knockout of WDR91 (KO-91) or WDR81 (KO-81) increases the sizes of early endosomes in HeLa cells in a Beclin1-dependent manner. Cells were imaged 24 h after transfection with 2xFYVE-mCherry expression vector ± siBeclin1. Bars, 5 µm. (C) Colocalization (arrows) of WDR81 or WDR91 with Beclin1 in immunostained cells. Insets show magnified (1.8×) views of boxed areas. Bars, 5 µm. (D–G) Co-IP of HA-Beclin1 with Flag-WDR81 (D), HA-Beclin1 with Myc-WDR91 (E), Flag-Vps34 with Myc-WDR81 (F), and Flag-Vps34 with Myc-WDR91 (G). Flag or Myc antibodies were used for IPs. Precipitated proteins were detected with HA, Myc, or Flag antibodies.
WDR91 and WDR81 suppress PI3K complex activity to regulate endosomal PtdIns3P levels
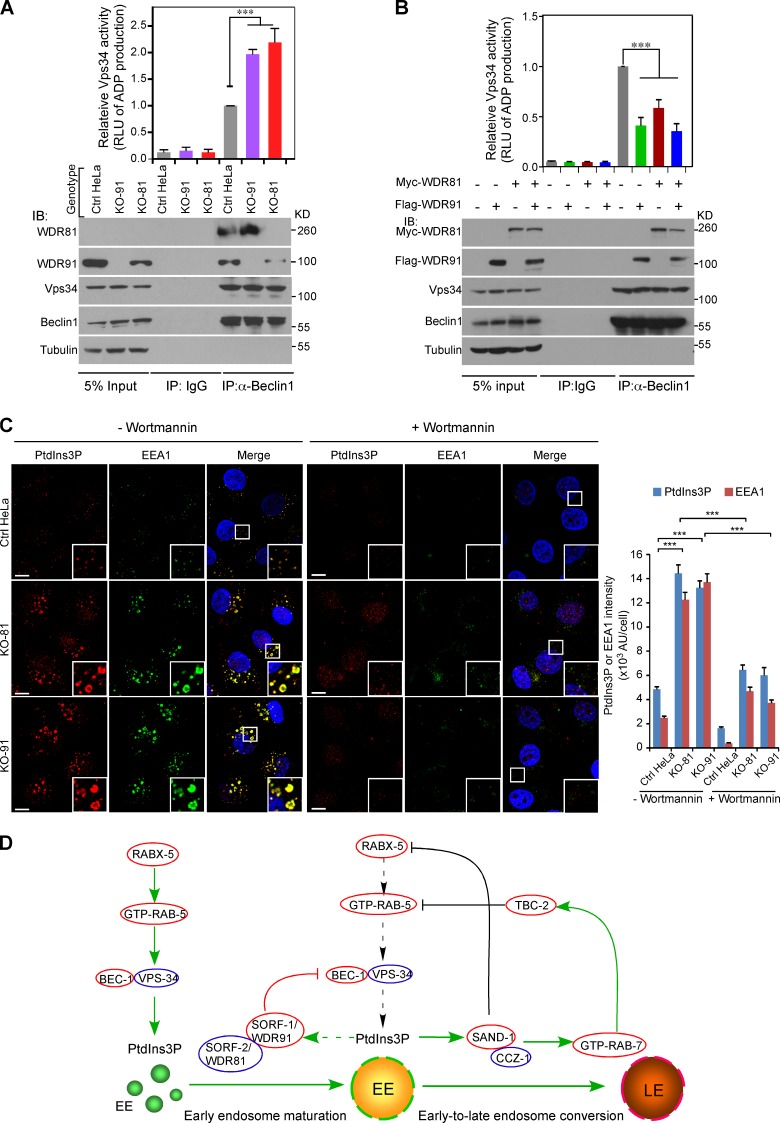
To understand the functional interaction of WDR91 and WDR81 with Beclin1, we immunoprecipitated Beclin1 from cell lysates of wild-type (control), WDR91 KO, and WDR81 KO HeLa cells and measured the activity of the PI3K complex. In control HeLa cells, WDR81, WDR91, and Vps34 were coprecipitated with Beclin1 (Fig. 10 A). In the absence of WDR81 or WDR91, the PI3K complex containing Beclin1 and Vps34 was much more active than the complex with WDR81 and WDR91 (Fig. 10 A). In contrast, reinforced expression of Myc-WDR81 or Flag-WDR91, or both, strongly suppressed the activity of the PI3K complex in the same kinase assays (Fig. 10 B). Thus, the interaction of WDR91 and WDR81 with Beclin1 inhibits the activity of the PI3K complex.
Figure 10.
WDR91 and WDR81 inhibit PI3K complex activity. (A) Loss of WDR91 or WDR81 enhances activity of the PI3K complex. Beclin1 was immunoprecipitated from cell lysates of control (Ctrl), KO-91, and KO-81 HeLa cells. Precipitated proteins were detected with indicated antibodies (bottom panel). Equal amounts of precipitated proteins from each genotype were examined for PI3K activity by measuring relative light unit of luminescence (RLU) of ADP converted from ATP. Data representing mean ± SEM are from three independent experiments and are normalized to PI3K activity in Ctrl cells (top panel). ***, P < 0.001. (B) Reinforced expression of Flag-WDR91 or/and Myc-WDR81 inhibits PI3K complex activity. HeLa cells were transfected with Flag-WDR91 or Myc-WDR81 expression vectors, or both. 48 h later, cell lysates were subjected to IP of Beclin1. Precipitates were examined with antibodies of indicated proteins (bottom panel) then analyzed for PI3K activity as in A. Data representing mean ± SEM are from three independent experiments and are normalized to PI3K activity in Ctrl cells (top panel). ***, P < 0.001. (C) Coimmunostaining of PtdIns3P with EEA1 in Ctrl, KO-91, and KO-81 HeLa cells treated without or with wortmannin (200 nM, 1 h). Insets show enlarged (2.5×) views of boxes in merged images. Bars, 10 µm. Right panel: PtdIns3P and EEA1 intensity was quantified in ≥100 cells. ***, P < 0.001. (D) Negative regulation of endosomal PtdIns3P levels by SORF-1/WDR91-SORF-2/WDR81 and Rab switch factors. Solid green arrows, activation; dashed green arrow, potential activation effect; black and red lines, inhibitory effects; dashed black arrows, termination of activation. EE, early endosome; LE, late endosome. See also Discussion.
We next examined endosomal PtdIns3P levels by immunostaining with a PtdIns3P antibody. PtdIns3P signals were much higher in WDR91 or WDR81 KO cells than in control cells (Fig. 10 C). Remarkably, the enhanced PtdIns3P signals significantly overlapped with the increased EEA1 signals in WDR91 or WDR81 KO cells, indicating that loss of WDR91 or WDR81 function indeed leads to elevated endosomal PtdIns3P (Fig. 10 C). Nevertheless, this effect was strongly inhibited by the PI3K inhibitor wortmannin (Fig. 10 C), confirming that WDR91 and WDR81 function through the PI3K complex. These findings suggest that WDR91 and WDR81 inhibit PI3K complex activity by interacting with Beclin1, thereby negatively regulating endosomal PtdIns3P levels.
Discussion
In this study, we identified two new factors, SORF-1 and SORF-2, that regulate endosomal PtdIns3P in C. elegans. Loss of sorf-1 or sorf-2 leads to elevation and prolonged existence of endosomal PtdIns3P, which induces excessive fusion of early endosomes. As a result, early-to-late endosome conversion and endosomal transport are delayed. Our results further suggest that negative regulation of endosomal PtdIns3P by SORF-1 and SORF-2 is likely achieved by their direct interaction with the BEC-1/Beclin1 subunit of the class III PI3K complex, though other mechanisms might also be involved (Fig. 10 D).
PtdIns3P can drive fusion of early endosomes through its effectors (Subramanian et al., 2010). During early-to-late endosome switching, a protein interaction cascade consisting of RABX-5, SAND-1, CCZ-1, RAB-7, and TBC-2 suppresses RAB-5–mediated PtdIns3P synthesis, because the “active” GTP-RAB-5 is responsible for recruitment of the PI3K complex to early endosomes (Chotard et al., 2010; Poteryaev et al., 2010; Pfeffer, 2013). SAND-1 senses the PtdIns3P level on early endosomes and displaces RABX-5 from endosome membranes; in the meantime, SAND-1 and CCZ-1 form a complex that converts RAB-7 into a GTP-bound “active” form (Nordmann et al., 2010; Poteryaev et al., 2010). GTP-RAB-7 further recruits TBC-2, the RAB-5 GAP, to inactivate RAB-5 on endosomes (Chotard et al., 2010). Thus, deletion of sand-1, ccz-1, rab-7, or tbc-2 leads to enhanced RAB-5 activity, which in turn increases endosomal PtdIns3P levels. Intriguingly, in C. elegans coelomocytes, endosomal PtdIns3P still turns over in the absence of sand-1, rab-7, or tbc-2, suggesting that alternative mechanisms exist that either inhibit PtdIns3P synthesis or promote its degradation on endosomes. Our findings reveal a mechanism by which endosomal PtdIns3P levels are properly controlled (Fig. 10 D). In this model, SORF-1 likely senses and binds to endosomal PtdIns3P and forms a complex with SORF-2. This complex interacts with BEC-1/Beclin1 and suppresses the activity of the PI3K complex, which acts in coordination with the protein interaction cascade (SAND-1–CCZ-1–RAB-7–TBC-2) that suppresses RAB-5 and hence RAB-5–mediated PI3K activity; thus, PtdIns3P synthesis on early endosomes is strongly inhibited (Fig. 10 D). On the other hand, endosomal PtdIns3P may be dephosphorylated by myotubularin family phosphatases, converted into PtdIns(3,5)P2 by PIKfyve/Fab1, or invaginated into the endosomal lumen for degradation (Schink et al., 2013), allowing early-to-late endosome conversion and endolysosomal cargo transport.
Our data also suggest that elevated PtdIns3P levels can partially rescue the endosome fusion defect caused by partial loss of function of the CORVET/HOPS complex. Studies in yeast and mammalian cells suggest that CORVET and HOPS serve as tethering complexes to mediate Rab5/Vps21-dependent early endosome fusion and Rab7/Ypt7-mediated late endosome fusion, respectively (Peplowska et al., 2007; Balderhaar et al., 2013; Pols et al., 2013). Deletion mutants of C. elegans HOPS complex genes show defective fusion of both endosomes and lysosomes, suggesting that the complex mediates both RAB-5– and RAB-7–dependent membrane fusion (Xiao et al., 2009). In this study, we found that loss of sorf-1 and sorf-2 partially rescued defective endosome fusion in deletion mutants of vps-11, -18, -39, and -41, suggesting that the enhanced PtdIns3P level can compensate for the functional loss of these VPS subunits. One explanation for the compensatory effect of PtdIns3P is that the elevated level of PtdIns3P likely promotes membrane fusion through other PtdIns3P effectors, for example, RABS-5/Rabenosyn-5 and EEA-1/EEA1, which play nonredundant roles to drive endosome fusion by facilitating formation of SNARE complexes on two opposing membranes (McBride et al., 1999; Nielsen et al., 2000; Ohya et al., 2009; Subramanian et al., 2010).
The negative regulation of endosomal PtdIns3P as revealed in C. elegans is probably conserved in mammals, because knockout of WDR91 or WDR81 similarly results in Beclin1-dependent enlargement and PtdIns3P enrichment of endosomes, which impairs lysosome-mediated EGFR degradation. In addition, WDR91 and WDR81 interact with one another, and both of them interact directly with Beclin1 and inhibit PI3K complex activity, similar to the situation in C. elegans (Fig. 10 D). WDR91 contains an extra WD40-repeat region compared with SORF-1, so other endosomal factors may interact with WDR91 through its WD40-repeat domain to regulate the activity of the WDR91-WDR81 complex. Our findings provide important clues for understanding the physiological functions of WDR91 and WDR81, and especially the role of WDR81 in developmental disorders such as cerebellar ataxia, mental retardation, and quadrupedal locomotion syndrome-2 (Gulsuner et al., 2011). Given that WDR81 and WDR91 interact with Beclin1, it will be of interest to investigate whether WDR81 or WDR91 are involved in autophagy-related cellular events. It will be very interesting to investigate whether WDR91 mutation is involved in endosome/lysosome-related human diseases.
Materials and methods
C. elegans strains and genetics
The Bristol strain N2 was used as wild type. sorf-2(yq4) and ppk-3(yq24) mutants were obtained by ethyl methanesulfonate (EMS) mutagenesis. sorf-1(tm3855) and sorf-2(tm5210) deletion mutants were isolated with trimethylpsoralen and UV mutagenesis and PCR screening. The ppk-3(yq24) allele has a S1448L mutation in the kinase domain of PPK-3. Other mutant alleles used in this study are listed by linkage groups: LG I, mtm-1(gk347186), C34B7.2(tm5202); LGII, rab-7(ok511), vps-11(ok1664), vps-18(tm1125), tbc-2(tm2241); LGIII, mtm-3(tm4475), vps-33.1(tm327), vps-16(ok719); LGIV, sand-1(ok1963), bec-1(ok700); LGV, unc-76(e911), mtm-9(ok3523), vps-39(tm2253); and LGX, vps-41(ok3433), mtm-5(ok469), mtm-6(ok330), ppk-3(n2668), piki-1(tm3171), Y34B4A.2(gk280145). Sterile mutants including vps-11, -16, -33, -39, and -41, rab-7, sand-1, and bec-1 deletion mutants were maintained as heterozygotes, and their phenotypes were examined in adult progeny from heterozygous mothers. The integrated arrays cdIs85(Punc-1222ᵡfyve::gfp), cdIs97(Punc-122mCherry::cup-5), cdIs131(Punc-122gfp::rab-5), bIs34(Prme-8gfp::rme-8), pwIs50(Plmp-1lmp-1::gfp), and pwIs126(Peea-1gfp::eea-1) were provided by H. Fares (University of Arizona, Tucson, AZ) and B. Grant (Rutgers University, Piscataway, NJ). The integrated array opIs334 (Pced-1yfp::2ᵡfyve) was provided by K.S. Ravichandran (University of Virginia, Charlottesville, VA). The integrated array tmIs225 (Pasp-1asp-1::dsRed) was provided by T. Katada (University of Tokyo, Bunkyo-ku, Tokyo, Japan). The integrated array qxIs605 (Pvps-34Flag::vps34) was provided by X. Wang (National Institute of Biological Sciences, Beijing, China). Other strains used in this study carrying integrated or extrachromosomal arrays are as follows: yqIs140 (Phsprab-7(T23N)), yqEx811 (Pctns-1mCherry::rab-7), yqEx723 (Punc-122cfp::2xFYVE; Pctns-1yfp::bec-1), yqEx815 (Psorf-1Flag::sorf-1), yqEx818 (Py71h2am.2Myc::sorf-2), yqEx980 (Pctns-1rme-8::mCherry), yqEx770 (Psorf-1gfp::sorf-1), and yqEx422 (Pctns-1gfp::sorf-2). The WormBase IDs of mutants and transgenic arrays are summarized in Table S1. Animals carrying the integrated array were outcrossed with the N2 strain four times. Deletion strains were outcrossed with the N2 strain at least four times. C. elegans cultures and genetic crosses were performed according to standard procedures.
RNAi and genome-wide RNAi screen
C. elegans RNAi experiments were performed using bacteria feeding assays as described previously (Chen et al., 2013). In brief, synchronized L4-stage animals were transferred to plates seeded with bacteria expressing dsRNA and cultured at 20°C. F1 progeny were observed under DIC microscopes when they grew to the age of 36–48 h after the L4 molt. With vps-18(tm1125) mutants, we screened the RNAi clones corresponding to genes on chromosomes X and V in the C. elegans RNAi library (generated by the Ahringer laboratory) and found that RNAi of the ZK563.5 gene increased the sizes of endocytic vesicles in the coelomocytes. RNAi treatment of vps-11(ok1664), -16(ok719), -33.1(tm327), -39(tm2253), and -41(ok3433) was performed by feeding the heterozygous L3–L4 animals with bacteria expressing dsRNA, and phenotypes were examined in adult progeny.
EMS screen and gene cloning
To screen for mutants suppressing the phenotypes of vps-18(tm1125) mutants, synchronized L4-stage vps-18(tm1125) animals were treated with 50 mM EMS for 4 h. The F2 progeny of EMS-treated animals were grown to the age of 24–48 h after the L4 molt at 20°C and observed under DIC microscopes. This allowed us to obtain suppressors showing larger organelles in coelomocytes than in vps-18 single mutants. A mutant, yq4, was obtained from a screen of 2,000 haploid genomes. Mapping of the yq4 mutation was performed in the vps-18(tm1125) background using single nucleotide polymorphism–based mapping as described previously (Davis et al., 2005), with minor modifications. As a result, yq4 was mapped to linkage group III (LG III), and a point mutation (AG to AA) was identified in the splice acceptor site immediately before the last exon of the F52C9.1 gene. The splice site was shifted 13 bp downstream of the original AG site, resulting in a frameshift in the remaining sequence (Fig. S1 C). A deletion mutant of F52C9.1, tm5210, was further generated, which contains a deletion of 1198 bp removing exons 3–5 and part of exon 2 (Fig. S1 C). yq4 and tm5210 failed to complement one another, indicating that their rescuing effect on the fusion defects in vps-18(tm1125) animals indeed resulted from loss of F52C9.1.
Microscopy and trafficking experiments in coelomocytes
For C. elegans imaging, adult animals were immobilized with 2.5 mM levamisole in M9 (1 l contains 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, and 1 mM MgSO4) and mounted on 2% agarose pads for imaging. DIC pictures were captured using an Axioimager M1 (100× 1.3-NA oil objective; Carl Zeiss) coupled with an AxioCam monochrome digital camera and Axiovision release 4.7 software. Fluorescence images were obtained with an inverted FV1000 confocal microscope system (IX81; Olympus) using a 60× 1.42-NA oil objective. Excitation was achieved using solid-state 488-nm and gas-state 595-nm lasers. All images were taken at 25°C. For measurement of fluorescence intensity, a line was drawn across the center of the endosome, and the mean gray values of membrane boundaries were measured using FV10-ASW 4.0a Viewer software (Olympus).
TR-BSA trafficking assays in C. elegans coelomocytes were performed as described by Zhang et al. (2001). In brief, TR-BSA (Sigma-Aldrich; 1 mg/ml in water) was injected into the body cavity of adult animals, which were then cultured on NGM plates seeded with Escherichia coli OP50 at 20°C. Animals were imaged by confocal microscopy at different time points after injection. For each time point, similar results were obtained in more than 10 coelomocytes from six different animals.
LysoTracker red staining of lysosomes in coelomocytes was performed by injecting LysoTracker red DND-99 (Invitrogen) at 0.4 mM in M9 buffer into the body cavity of adult animals. After recovery for 4 h at 20°C, the organelles stained with LysoTracker red in the coelomocytes were examined under a confocal microscope with gas-state 595-nm lasers.
Expression vectors
pCMV-Flag-BEC-1, pmCherry-N1-Beclin1, and pCMV-Flag-Vps34 were provided by H. Zhang (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). pGEX4T1-BEC-1, pET28a-VPS-34, pCMV-Flag-VPS-34, and pCMV-HA-Beclin1 were provided by X. Wang. Other C. elegans expression vectors are listed in Table S2. Bacterial and mammalian cell expression vectors are listed in Table S3.
Mammalian cell culture and small RNAi
HeLa or HEK293 cells were cultured at 37°C with 5% CO2 in DMEM supplemented with 10% FBS (HyClone), 100 U/ml penicillin, and 100 mg/ml streptomycin. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
siRNA was achieved by transfection of a Beclin1-specific RNA oligo: 5′-CAGUUUGGCACAAUCAAUA-3′ (Zhong et al., 2009). The oligo used for control siRNA is 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were harvested for further analysis 48 h after transfection.
Generation of WDR91 and WDR81 knockout HeLa cells
To generate WDR81-knockout cells, two WDR81 guide RNAs, 81sgRNA1 (5′-TCAGGTCCCTCTCCACGCTC-3′) and 81sgRNA2 (5′-CGATCCCAGGCAGCTGGCTC-3′), were individually cloned into the pLKO-GFP vector, which was a gift from Z. Shen (National Institute of Biological Sciences, Beijing, China), and cotransfected with the D10A Cas9 mutant into HeLa cells. 72 h later, GFP-positive cells were sorted by FACS and cultured for 10 d. 48 colonies were picked and examined for deletion in the WDR81 gene by PCR, and the deletion was further confirmed by sequencing. Similar strategies were applied to generate WDR91-knockout cells using two WDR91 guide RNAs, 91sgRNA1 (5′-CGCGGAAGAGCAGGTACTCC-3′) and 91sgRNA2 (5′-GCAGCTGGACGCCGAGATCA-3′).
Time-lapse imaging
Time-lapse imaging in C. elegans was performed at 20°C under a 100× 1.40-NA oil objective using the DeltaVision imaging system (DV Elite; GE Healthcare) coupled with a CoolSNAP camera (Photometric Scientific). Animals (n < 5) were anaesthetized in 2 µl M9 buffer containing 1 mM levamisole and covered with a 2% agar pad in a glass-bottom dish (MatTek). The humidity in the dish was maintained with a piece of tissue paper saturated with water. Animals were rescued to examine their viability after imaging. Images of coelomocytes in each genotype were captured every 30 s for 120 min, with a Z-series of 0.5 µm/section for a total of 30 sections for each time point. Images were deconvoluted with the Enhanced Ratio option, and the Z-series of the whole coelomocyte were projected to form one image using the maximum-intensity projection option in softWoRx software coupled with DV Elite. The number of early endosomes that disappeared per hour, the duration of the 2xFYVE signal on endosomes, and the diameters of endosomes or lysosomes were measured manually based on the projected image using softWoRx. To record the dynamic change of 2xFYVE::GFP-labeled endosomes induced by RAB-7(T23N), transgenic animals containing Phsp::rab-7(T23N) were cultured at 33°C for 30 min and then at 20°C for 15 min before imaging. To record the dynamics of endosomes in HeLa cells, cells were grown at 37°C in DMEM supplemented with 10% FBS in glass-bottom dishes placed in a humidified chamber (Chamlide) supplemented with 5% CO2. Images were captured every 30 s for 120 min with a Z-series of 0.4 µm/section for a total of 20 sections using the DeltaVision imaging system. Images were similarly deconvoluted and projected. The excitation filters used for GFP and mCherry in all images were 488 and 559 nm, respectively.
Recombinant proteins and GST pull-down
Recombinant GST-BEC-1 and MBP-SORF-1 proteins were expressed in BL21(DE3) bacterial cells and purified with glutathione-Sepharose beads (GE Healthcare) according to the instructions provided by the supplier. 35S-labeled VPS-34, SORF-1, or SORF-2 proteins were prepared by in vitro translation. Purified GST or GST-BEC-1 proteins (2.5 µg each) immobilized on glutathione-Sepharose beads or amylose resin were incubated with 35S-labeled VPS-34, SORF-1, or SORF-2 at 4°C for 4 h and then washed extensively. Bound proteins were resolved on SDS-PAGE and visualized by autoradiography.
Antibodies and reagents
Beclin1 rabbit polyclonal antibody was purchased from Medical & Biological Laboratories; EEA1 and RAB5 mouse monoclonal antibodies from BD Bioscience; EEA1, Vps34, Beclin1, and Lamp1 rabbit monoclonal antibodies from Cell Signaling Technologies; EGFR rabbit polyclonal antibody from Abcam; PtdIns3P mouse monoclonal antibody from Echelon Biosciences; and Flag and Myc mouse monoclonal antibodies and MBP rabbit polyclonal antibody from Sigma-Aldrich. LMP-1 mouse polyclonal antibody was a gift from X. Wang. WDR81 antibody was generated in guinea pigs and rabbits by injecting purified GST-WDR81(332-604). WDR91 antibody was generated in mouse by injecting GST-WDR91(406-730). BEC-1 antibody was generated in rabbits or mice by injecting purified GST-BEC-1. GFP antibody was generated in mouse and rabbits by injecting purified His-GFP. Alexa Fluor dye–conjugated secondary antibodies, LysoTracker red DND-99, Dextran blue (MW 10,000), and Alexa Fluor 647–conjugated EGF were purchased from Life Technologies; TR-BSA from Sigma-Aldrich; ADP-GloTM Kinase Assay kit from Promega; and lipid strips from Echelon Biosciences.
Immunoprecipitations
For immunoprecipitation, cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, and 1 mM PMSF), and immunoprecipitations were performed as described (Chen et al., 2013) using individual antibodies. Precipitated proteins were resolved by SDS-PAGE and detected by Western blot using different antibodies (see Antibodies and reagents).
Immunostaining and imaging of mammalian cells
Cells grown on coverslips were fixed in 4% paraformaldehyde followed by permeabilization with 0.2% saponin for 8 min. After extensive washing with PBS, coverslips were incubated in blocking buffer 1 (5% BSA and PBS) for 1 h at RT and then incubated with primary antibodies in the same buffer at 4°C overnight. Cells were washed extensively again and incubated with cyanine 3– or FITC-conjugated secondary antibodies for 1 h at RT. After another round of thorough washing, cells were sealed with VectaShield mounting medium (Vector Laboratories) for confocal microscopy analysis.
For immunostaining of endogenous PtdIns3P, a detergent-free method was used with modifications (Munson et al., 2015). In brief, cells grown on coverslips were washed with PBS and ice-cold glutamate buffer (25 mM Hepes, pH 7.4, 25 mM KCl, 2.5 mM magnesium-acetate, 5 mM EGTA, and 150 mM potassium-glutamate) then frozen in liquid N2 until the bubbling stopped. Once thawed, cells were washed twice with ice-cold glutamate buffer. Cells were then fixed with 3.7% (wt/vol) formaldehyde in 200 mM Hepes, pH 7.4, for 30 min at RT. Formaldehyde was removed by washing twice with 10 mM Hepes, pH 7.4, in DMEM, followed by washing twice with blocking buffer 2 (1% BSA and PBS; 10 min for each). The coverslips were subsequently incubated in blocking buffer 2 for an additional 30 min, and then incubated with PtdIns3P antibody (5 µg/ml) and EEA1 antibody in the same buffer at 4°C overnight. Cells were washed extensively again with blocking buffer 2 and incubated with cyanine 3– and FITC-conjugated secondary antibodies for 1 h at RT. After another round of thorough washing, coverslips were washed once more with double-distilled H2O and mounted with VectaShield mounting medium for confocal microscopy analysis.
Lipid-binding assay
Membrane strips (Echelon Biosciences) were blocked with 1% nonfat dried milk in PBS for 1 h at RT and then incubated overnight at 4°C with 2.5 µg/ml MBP-fused proteins in 1% nonfat dried milk in PBS. Membrane strips were washed and blotted with MBP antibody (Sigma-Aldrich).
In vitro PI3K complex activity assay
HeLa cells of different genotype or transfected with the indicated vectors for 48 h were harvested and lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail). Endogenous Vps34 were immunoprecipitated by antibody against Beclin1 (MBL). Immunoprecipitated beads were extensively washed with lysis buffer and further washed twice with reaction buffer (40 mM Tris-HCl, pH 7.5, 20 mM MgCl2, and 1 mg/ml BSA). The beads were then incubated with 10 µg sonicated phosphatidylinositol (Sigma-Aldrich) and 1 µl ATP (10 mM) in 30 µl reaction buffer for 30 min at RT. Conversion of ATP to ADP was measured with an ADP-GloTM kinase assay kit following the instructions provided by the manufacturer.
To examine the activity of the C. elegans PI3K complex, adult animals expressing Flag::VPS-34 were harvested and frozen in liquid N2 with lysis buffer. The animals were then homogenized completely with tissue grinders. Endogenous BEC-1 was immunoprecipitated with BEC-1 antibody, and coprecipitated VPS-34 proteins were examined by Western blot with Flag antibody (Sigma-Aldrich). PI3K activity was measured as described previously.
Phase separation of membrane proteins
Triton X-114 phase separation was performed as described previously (Bordier, 1981). In brief, worms expressing Flag::SORF-1 and Myc::SORF-2 or HeLa cells expressing Myc-WDR81 or Myc-WDR91 were lysed in 0.5 ml Triton X-114 buffer (Tris-HCl, pH 7.5, 2% Triton X-114, 100 mM NaCl, and 1 mM PMSF) for 30 min on ice, and the nuclei and cell debris were removed by centrifugation at 10,000 g at 4°C for 10 min. The proteins were then partitioned into detergent and aqueous phases by incubation at 37°C for 15 min and centrifugation at 1,800 g for 5 min. Proteins in each fraction were harvested by acetone precipitation and analyzed by immunoblotting.
Statistical analysis
Data were analyzed with Prism (GraphPad Software) or Excel (Microsoft Office) to generate curves or bar graphics. Error bars represent SEM. The two-tailed unpaired t test was used for statistical analysis of two groups of samples. One-way analysis of variance with a Newman–Keuls posttest was used to evaluate statistical significance of multiple groups of samples. P > 0.05 was considered not significant.
Online supplemental material
Fig. S1 describes the identification of sorf-1 and sorf-2 in C. elegans. Fig. S2 analyzes the dynamic change of GFP::RAB-5 and mCherry::RAB-7 on endosomes in coelomocytes in N2, sorf-1(tm3855), and sorf-2(tm5210) animals. Fig. S3 characterizes the giant endosomes induced by loss of rab-7 and sorf-1 or sorf-2. Fig. S4 characterizes the genetic interaction of sorf-1 or sorf-2 with genes involved in endosomal PtdIns3P metabolism. Fig. S5 shows the interaction of SORF-1 or SORF-2 with BEC-1, VPS-34, RAB-5, and RAB-7 and the role of human WDR91 and WDR81 in macropinocytosis. Video 1 shows that loss of sorf-1 or sorf-2 delays early-to-late endosome conversion. Video 2 shows that loss of sorf-1 or sorf-2 delays endosomal RAB-5-to-RAB-7 switch in coelomocytes. Videos 3 and 4 show that endosomal PtdIns3P turnover is abrogated in the double mutants of sorf-1 or sorf-2 with rab-7 or sand-1, respectively. Video 5 shows that the giant endosomes in the double mutants of sorf-1 or sorf-2 with Rab switch genes result from excessive fusion of early endosomes. Video 6 shows that GFP-WDR81 and GFP-WDR91 appear on early endosomes with progressive disappearance of 2xFYVE-mCherry. Table S1 summarizes the WormBase IDs of mutants and transgenic arrays. Tables S2 and S3 list C. elegans, bacterial, and mammalian expression vectors. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506081/DC1.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetic Center and Drs. X.C. Wang, H. Zhang, H. Fares, B. Grant, T. Katada, A. Spang, and K.S. Ravichandran for strains and reagents, Drs. X.C. Wang and H. Zhang for constructive suggestions, Drs. M.J. Munson and I.G. Ganley for technical assistance, and Dr. I. Hanson for manuscript proofreading.
This research was supported by grants from National Science Foundation of China (31230043 and 31025015), the National Basic Research Program of China (2013CB910102 and 2011CB910102), and the Chinese Academy of Sciences (KJZD-EW-L08).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CORVET
- class C core vacuole/endosome tethering
- DIC
- differential interference contrast
- EGFR
- EGF receptor
- EMS
- ethyl methanesulfonate
- GAP
- GTPase-activating protein
- HOPS
- homotypic fusion and vacuole protein sorting
- KO
- knockout
- MBP
- maltose-binding protein
- PI3K
- phosphatidylinositol 3-kinase
- PtdIns3P
- phosphatidylinositol 3-phosphate
- TR-BSA
- Texas red–BSA
References
- Balderhaar H.J., Lachmann J., Yavavli E., Bröcker C., Lürick A., and Ungermann C.. 2013. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc. Natl. Acad. Sci. USA. 110:3823–3828. 10.1073/pnas.1221785110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., and Grant B.D.. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073. 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604–1607. [PubMed] [Google Scholar]
- Cao C., Laporte J., Backer J.M., Wandinger-Ness A., and Stein M.P.. 2007. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 8:1052–1067. 10.1111/j.1600-0854.2007.00586.x [DOI] [PubMed] [Google Scholar]
- Cao C., Backer J.M., Laporte J., Bedrick E.J., and Wandinger-Ness A.. 2008. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol. Biol. Cell. 19:3334–3346. 10.1091/mbc.E08-04-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Jian Y., Liu X., Zhang Y., Liang J., Qi X., Du H., Zou W., Chen L., Chai Y., et al. . 2013. Clathrin and AP2 are required for phagocytic receptor-mediated apoptotic cell clearance in Caenorhabditis elegans. PLoS Genet. 9:e1003517 10.1371/journal.pgen.1003517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Mishra A.K., Sylvain M.A., Tuck S., Lambright D.G., and Rocheleau C.E.. 2010. TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 21:2285–2296. 10.1091/mbc.E09-11-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., McBride H.M., Burgoyne R.D., and Zerial M.. 1999a The Rab5 effector EEA1 is a core component of endosome docking. Nature. 397:621–625. 10.1038/17618 [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.C., Waterfield M.D., Backer J.M., and Zerial M.. 1999b Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1:249–252. 10.1038/12075 [DOI] [PubMed] [Google Scholar]
- Davis M.W., Hammarlund M., Harrach T., Hullett P., Olsen S., and Jorgensen E.M.. 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 6:118 10.1186/1471-2164-6-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Fares H., and Greenwald I.. 2001a Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 159:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., and Greenwald I.. 2001b Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 28:64–68. 10.1038/ng0501-64 [DOI] [PubMed] [Google Scholar]
- Funderburk S.F., Wang Q.J., and Yue Z.. 2010. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 20:355–362. 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter C.E., Collinson L.M., Backer J.M., and Hopkins C.R.. 2001. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 155:1251–1264. 10.1083/jcb.200108152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando K., Kuroyanagi H., Kobayashi T., Murate M., Fujimoto K., Okabe S., and Mitani S.. 2007. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 8:152–157. 10.1038/sj.embor.7400882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., and Stenmark H.. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19:4577–4588. 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.D., and Sato M.. 2006. Intracellular trafficking. WormBook. 2006:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S., Tekinay A.B., Doerschner K., Boyaci H., Bilguvar K., Unal H., Ors A., Onat O.E., Atalar E., Basak A.N., et al. . 2011. Homozygosity mapping and targeted genomic sequencing reveal the gene responsible for cerebellar hypoplasia and quadrupedal locomotion in a consanguineous kindred. Genome Res. 21:1995–2003. 10.1101/gr.126110.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Lippé R., McBride H.M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M., and Zerial M.. 1997. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 90:1149–1159. 10.1016/S0092-8674(00)80380-3 [DOI] [PubMed] [Google Scholar]
- Huotari J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M.C., and Teasdale R.D.. 2009. Defining macropinocytosis. Traffic. 10:364–371. 10.1111/j.1600-0854.2009.00878.x [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., and Wang X.. 2009. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 136:2445–2455. 10.1242/dev.035949 [DOI] [PubMed] [Google Scholar]
- Lippé R., Miaczynska M., Rybin V., Runge A., and Zerial M.. 2001. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol. Biol. Cell. 12:2219–2228. 10.1091/mbc.12.7.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., Lin L., Kovács A.L., Yu L., and Zhang H.. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell. 21:343–357. 10.1016/j.devcel.2011.06.024 [DOI] [PubMed] [Google Scholar]
- McBride H.M., Rybin V., Murphy C., Giner A., Teasdale R., and Zerial M.. 1999. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 98:377–386. 10.1016/S0092-8674(00)81966-2 [DOI] [PubMed] [Google Scholar]
- McKnight N.C., Zhong Y., Wold M.S., Gong S., Phillips G.R., Dou Z., Zhao Y., Heintz N., Zong W.X., and Yue Z.. 2014. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 10:e1004626 10.1371/journal.pgen.1004626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M.J., Allen G.F., Toth R., Campbell D.G., Lucocq J.M., and Ganley I.G.. 2015. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 34:2272–2290. 10.15252/embj.201590992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.T., Panaretou C., Stenmark H., Miaczynska M., and Backer J.M.. 2002. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 3:416–427. 10.1034/j.1600-0854.2002.30605.x [DOI] [PubMed] [Google Scholar]
- Nakae I., Fujino T., Kobayashi T., Sasaki A., Kikko Y., Fukuyama M., Gengyo-Ando K., Mitani S., Kontani K., and Katada T.. 2010. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol. Biol. Cell. 21:2434–2442. 10.1091/mbc.E09-12-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B., and Zerial M.. 2000. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151:601–612. 10.1083/jcb.151.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C., Almendinger J., Gysi S., Gómez-Orte E., Kaech A., Hengartner M.O., Schnabel R., Moreno S., and Cabello J.. 2010. ccz-1 mediates the digestion of apoptotic corpses in C. elegans. J. Cell Sci. 123:2001–2007. 10.1242/jcs.062331 [DOI] [PubMed] [Google Scholar]
- Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Ohya T., Miaczynska M., Coskun U., Lommer B., Runge A., Drechsel D., Kalaidzidis Y., and Zerial M.. 2009. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 459:1091–1097. 10.1038/nature08107 [DOI] [PubMed] [Google Scholar]
- Pedersen N.M., Raiborg C., Brech A., Skarpen E., Roxrud I., Platta H.W., Liestøl K., and Stenmark H.. 2012. The PtdIns3P-binding protein Phafin 2 mediates epidermal growth factor receptor degradation by promoting endosome fusion. Traffic. 13:1547–1563. 10.1111/j.1600-0854.2012.01400.x [DOI] [PubMed] [Google Scholar]
- Peplowska K., Markgraf D.F., Ostrowicz C.W., Bange G., and Ungermann C.. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell. 12:739–750. 10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. 2013. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25:414–419. 10.1016/j.ceb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemel R.L., Lobingier B.T., Brett C.L., Angers C.G., Nickerson D.P., Paulsel A., Sprague D., and Merz A.J.. 2011. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol. Biol. Cell. 22:1353–1363. 10.1091/mbc.E10-03-0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols M.S., ten Brink C., Gosavi P., Oorschot V., and Klumperman J.. 2013. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 14:219–232. 10.1111/tra.12027 [DOI] [PubMed] [Google Scholar]
- Poteryaev D., Fares H., Bowerman B., and Spang A.. 2007. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 26:301–312. 10.1038/sj.emboj.7601498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D., Datta S., Ackema K., Zerial M., and Spang A.. 2010. Identification of the switch in early-to-late endosome transition. Cell. 141:497–508. 10.1016/j.cell.2010.03.011 [DOI] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Ruck A., Attonito J., Garces K.T., Núnez L., Palmisano N.J., Rubel Z., Bai Z., Nguyen K.C., Sun L., Grant B.D., et al. . 2011. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 7:386–400. 10.4161/auto.7.4.14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Norris A., Sato M., and Grant B.D.. 2014. C. elegans as a model for membrane traffic. WormBook. 2014:1–47. 10.1895/wormbook.1.77.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schink K.O., Raiborg C., and Stenmark H.. 2013. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. BioEssays. 35:900–912. [DOI] [PubMed] [Google Scholar]
- Schnatwinkel C., Christoforidis S., Lindsay M.R., Uttenweiler-Joseph S., Wilm M., Parton R.G., and Zerial M.. 2004. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2:E261 10.1371/journal.pbio.0020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.N. 2012. The retromer complex - endosomal protein recycling and beyond. J. Cell Sci. 125:4693–4702. 10.1242/jcs.103440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M.P., Feng Y., Cooper K.L., Welford A.M., and Wandinger-Ness A.. 2003. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 4:754–771. 10.1034/j.1600-0854.2003.00133.x [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Subramanian D., Laketa V., Müller R., Tischer C., Zarbakhsh S., Pepperkok R., and Schultz C.. 2010. Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat. Chem. Biol. 6:324–326. 10.1038/nchembio.348 [DOI] [PubMed] [Google Scholar]
- Thoresen S.B., Pedersen N.M., Liestøl K., and Stenmark H.. 2010. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 316:3368–3378. 10.1016/j.yexcr.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Wickner W. 2010. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu. Rev. Cell Dev. Biol. 26:115–136. 10.1146/annurev-cellbio-100109-104131 [DOI] [PubMed] [Google Scholar]
- Wurmser A.E., and Emr S.D.. 1998. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 17:4930–4942. 10.1093/emboj/17.17.4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Chen D., Fang Z., Xu J., Sun X., Song S., Liu J., and Yang C.. 2009. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol. Biol. Cell. 20:21–32. 10.1091/mbc.E08-04-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Grant B., and Hirsh D.. 2001. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol. Biol. Cell. 12:2011–2021. 10.1091/mbc.12.7.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., and Yue Z.. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11:468–476. 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.