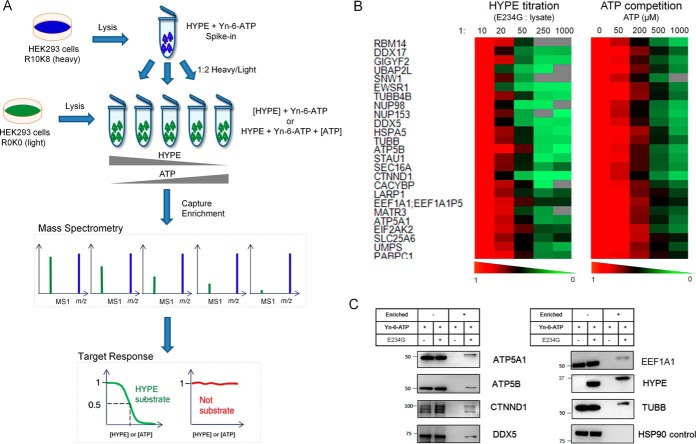
Fig. 2.
Validation of AMPylation candidates. (A) Schematic diagram of the spike-in SILAC strategy used for substrate validation in terms of dose-dependent HYPE and ATP enrichment specificity. (B) Heat maps of quantitative MS-based response of 25 high-confidence AMPylation substrates to HYPE titration or ATP competition. Color coding represents normalized levels (five conditions normalized to 1:10 w/w enzyme to lysate or 0 μm ATP) of substrate AMPylation in response to a range of HYPE and ATP concentrations. Protein substrates are reported by gene names. (C) Western blot validation of selected high-confidence substrates.