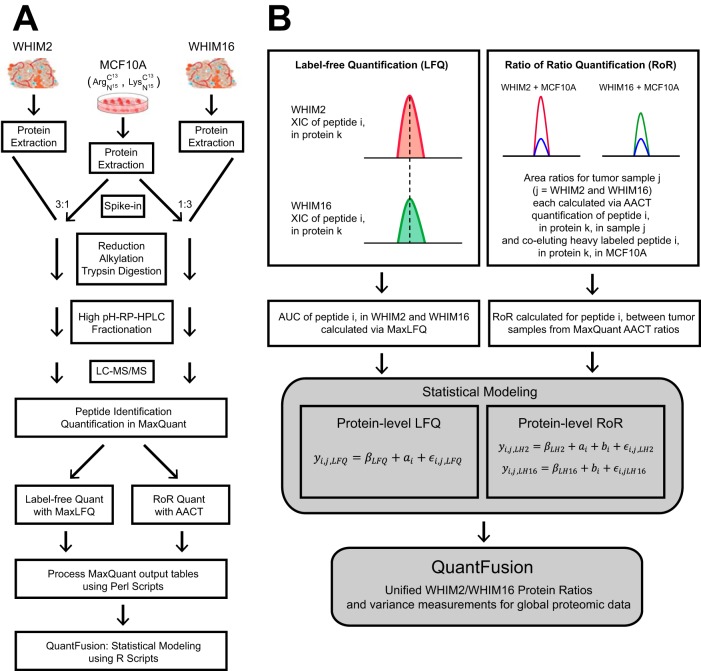
Fig. 1.
Proteomic workflow and QuantFusion model. A, proteomics workflow to analyze PDX tumors with heavy labeled added-in standard peptides derived from an MCF10A cell line. The cell line proteins were added to each tumor lysate at a protein mass ratio of 1:3, reduced, alkylated, and subjected to dual digestion with an endoproteinase Lys-C-trypsin enzyme combination. The resulting peptides were fractionated by high pH reversed phase liquid chromatography into 30 fractions that were non-contiguously recombined into five fractions and used for global LC-MS proteomic analysis. All data were subjected to LFQ analysis and AACT ratio determination. The LFQ and AACT outputs were processed using a Perl script and subjected to statistical modeling. B, graphical depiction of LFQ and RoR quantitation schemes and statistical model that merges LFQ and RoR estimates into a unified estimate.