Figure 1. Phylogenetic network analysis.
SplitsTree phylogenetic networks were constructed using (A) each isolate for the collection of T.b. brucei (Tbb), T.b. rhodesiense (Tbr), T.b. gambiense Group 1 (Tbg1) and T.b. gambiense Group 2 (Tbg2) and (B) for just T.b. gambiense Group 1 isolates. The number of samples in each group is indicated in parenthesis.
DOI: http://dx.doi.org/10.7554/eLife.11473.003
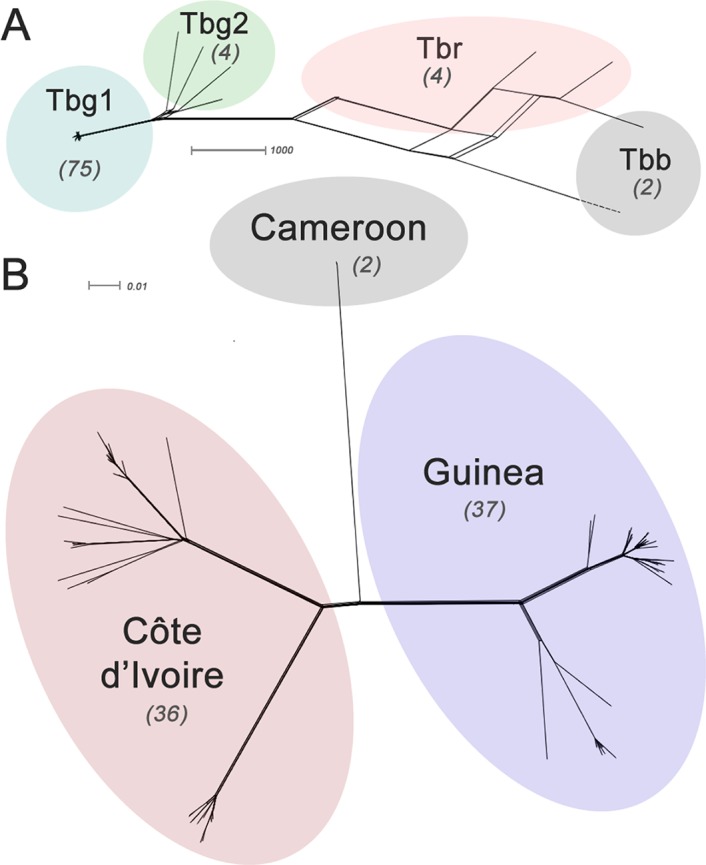
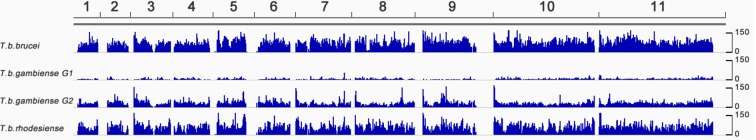
Figure 1—figure supplement 1. Genome-wide SNP density map for each sub-species.
Figure 1—figure supplement 2. Weir and Cockerham’s fis.
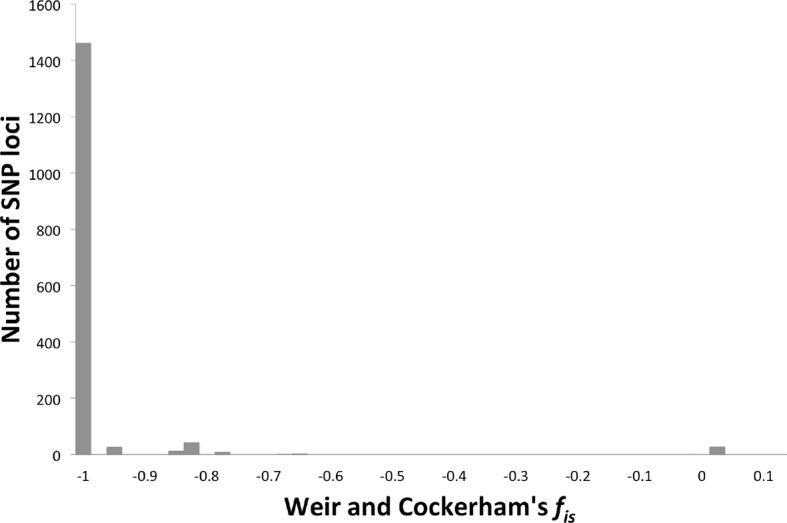
Figure 1—figure supplement 3. Genome-wide linkage disequilibrium (LD) among T.b. gambiense Group 1 parasites.