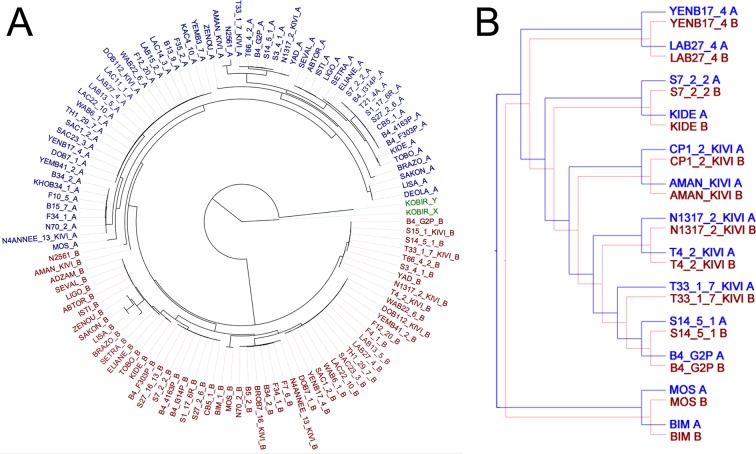
Figure 4. Haplotype co-evolution of chromosome 8.
(A) Phylogenetic tree of phased haplotype sequences of chromosome 8 shows a complete divergence between A (blue) and B (red) haplotypes of T.b. gambiense Group 1 (identical genotypes removed). The tree is rooted using a Group 2 isolate (green); (B) Co-phylogenetic analysis reveals 100% consensus between the A and B haplotype trees of a subset of T.b. gambiense Group 1 isolates.
DOI: http://dx.doi.org/10.7554/eLife.11473.016
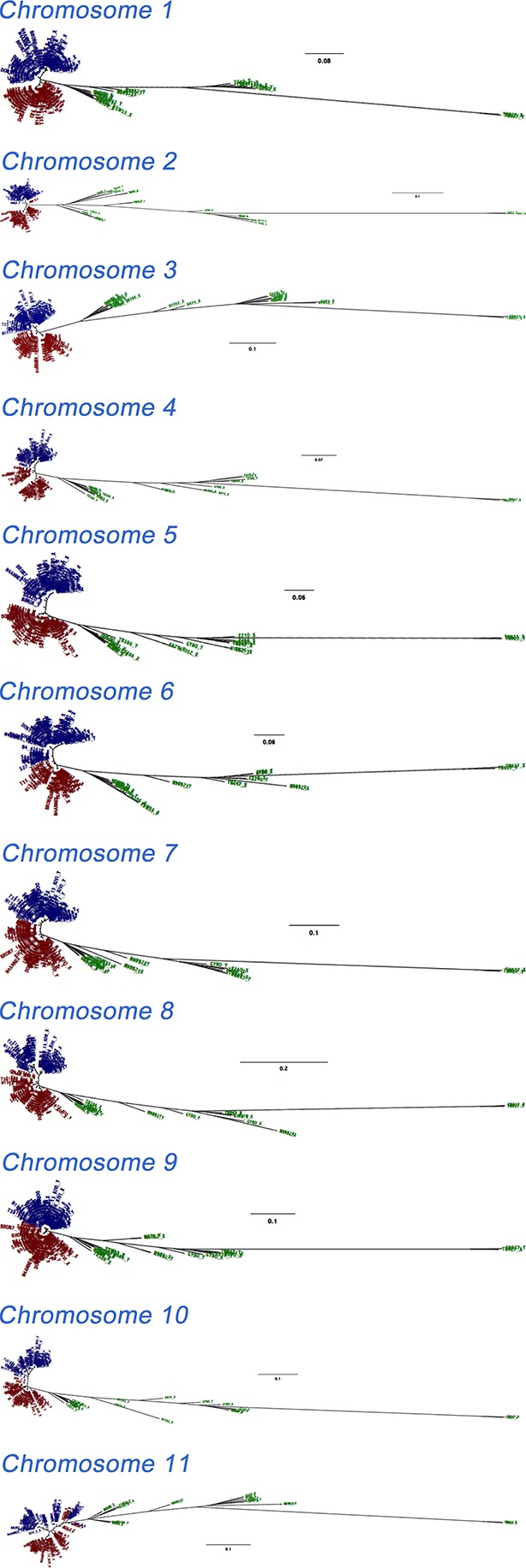
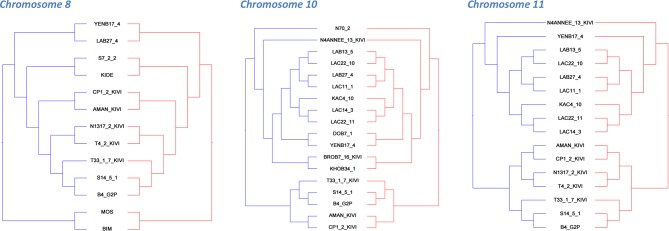
Figure 4—figure supplement 1. Phylogenetic trees of phased data showing ‘A’ and ‘B’ haplotypes.