Abstract
The enzyme activation-induced deaminase (AID) targets the immunoglobulin loci in activated B cells and creates DNA mutations in the antigen-binding variable region and DNA breaks in the switch region through processes known, respectively, as somatic hypermutation and class switch recombination. AID deaminates cytosine to uracil in DNA to create a U:G mismatch. During somatic hypermutation, the MutSα complex binds to the mismatch, and the error-prone DNA polymerase η generates mutations at A and T bases. During class switch recombination, both MutSα and MutLα complexes bind to the mismatch, resulting in double-strand break formation and end-joining. This review is centered on the mechanisms of how the MMR pathway is commandeered by B cells to generate antibody diversity.
Keywords: activation-induced deaminase, class switch recombination, DNA polymerase η, mismatch repair, somatic hypermutation
1. Introduction to AID and canonical DNA repair
Cells have evolved multiple pathways to maintain genomic integrity. These pathways include mismatch repair (MMR) to correct DNA replication errors, base excision repair (BER) and nucleotide excision repair to mend base damage from genotoxic agents, and translesion synthesis to bypass lesions. In most cells, these pathways work to efficiently remove DNA mispairs and damaged bases, and faithfully restore DNA to its original sequence. However, B cells use the MMR and BER pathways to generate DNA mutations as part of the antibody diversification process. Initially, the antibody repertoire is created in pre-B cells by the recombination of immunoglobulin (Ig) V(D)J (variable, diversity, joining) gene segments, and by the pairing of heavy and kappa or lambda light chains [1]. The antibody pool is subsequently expanded in mature B cells upon antigen exposure. These antigen-activated B cells undergo further diversification through somatic hypermutation (SHM) of rearranged variable genes and class switch recombination (CSR) of heavy chain constant genes, both of which are initiated by the enzyme activation-induced deaminase (AID) [2].
AID was discovered by Honjo and colleagues in 1999 [3], and was shown to be a member of the mRNA-editing APOBEC protein family [4]. However, further work by Neuberger and others revealed that AID acts upon DNA [5-7], where it deaminates cytosine to uracil in single-strand regions of DNA formed during transcription [8]. The protein is highly expressed in germinal centers from spleen, lymph nodes, and Peyer's patches [3, 9]. AID features an 11 amino acid C-terminal recognition loop, LYFCEDRKAEP, that favors binding to the C in WGCW (where W = A or T) sequence hotspots and deaminates both DNA strands [10-14]. The complete mechanism behind the upregulation and targeting of AID activity exclusively to the Ig loci is currently unknown, although enhancer regions and RNA polymerase II pausing are believed to play major roles [15-18]. AID targeting must be strictly regulated, because deaminations in non-Ig genes can generate translocations that lead to the development of diseases such as B-cell lymphomas [19]. The AID-induced U:G mismatch will mimic T:G, resulting in a C:G to T:A transition following DNA replication of the uracil. Alternatively, the improper uracil either can be repaired via canonical repair pathways, or can employ disrupted repair and translesion polymerases (pol) to generate antibody diversification by SHM and CSR.
As discussed in more detail elsewhere in this issue [20], DNA repair of base damages, including mismatched uracils, relies on the MMR and BER pathways [21, 22]. Canonical MMR uses a heterodimer complex formed by either MutSα, consisting of MSH2 and MSH6, or MutSβ, formed by MSH2 and MSH3, to recognize and bind to mismatches. MutSα targets single nucleotide mismatches, while MutSβ targets loops formed by inserts, deletions, and multi-base mispairs. A MutL heterodimer, containing either MLH1 and PMS2 (MutLα), MLH1 and PMS1 (MutLβ), or MLH1 and MLH3 (MutLγ) is then recruited to the mismatch. This review will emphasize MutLα, as the other MutL complexes may not be involved in the immune response [23]. MutLα introduces a nearby nick that acts as an exonuclease entry point. Exonuclease 1 (EXO1) removes the mismatch and adjacent bases, creating a single-strand gap. The PCNA sliding clamp recruits a high-fidelity DNA pol, such as pol δ or ε, to accurately resynthesize the gap, followed by DNA ligase I to seal the freshly-repaired strand. Alternatively, BER uses uracil DNA glycosylase (UNG) to remove rogue uracils. This leaves behind an abasic site, which is then cleaved by an apurinic/apyrimidinic endonuclease (APE), producing a single-strand break. Pol β excises the 5’ deoxyribose phosphate group and inserts the correct base, and DNA ligase III closes the nick.
Although canonical DNA repair is desirable under most circumstances, a significantly altered process ensues in B cells during antibody development. In a mechanism initially proposed by Neuberger [24], adjustments to the MMR pathway can introduce mutations at A and T bases, while a modified BER pathway is responsible for generating mutations at C and G bases. MMR and BER proteins also participate in switching between constant genes during CSR. This review focuses on how the MMR pathway is manipulated by B cells to generate antibody diversity.
2. MutSα complex generates A:T mutations during SHM
2.1. Pathways responsible for creating Ig variable region diversity
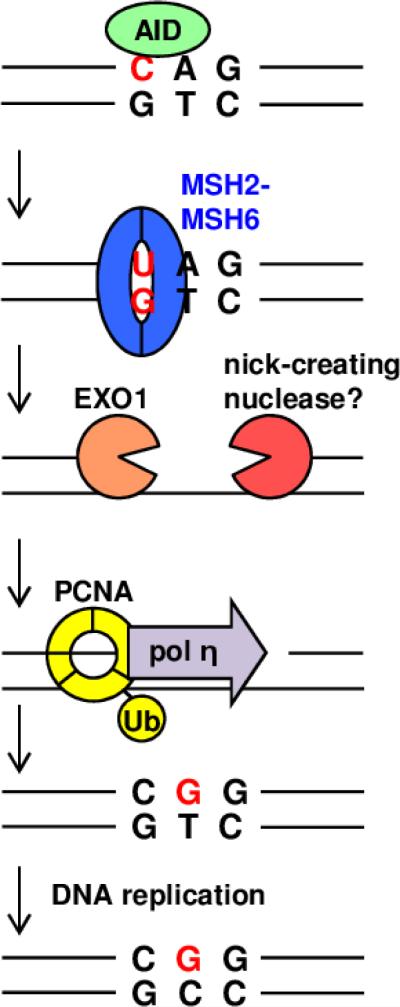
Antibodies contain both a rearranged V(D)J gene, which regulates antigen binding, and a constant gene, which determines isotype. Upon antigen exposure, B cells in germinal centers undergo successive rounds of SHM [25]. Mutations occur in two regions of DNA: (1) the variable region containing the rearranged VDJ or VJ gene, and (2) the switch region preceding each constant gene. Mutations start just downstream of transcription start sites in the promoter (variable region) and intronic enhancer (switch region), indicating that transcription is necessary for AID activity [26, 27]. These mutations typically occur as single base substitutions and form at an elevated frequency of 10−2 mutations/bp, compared to spontaneous mutation in other loci, which occurs at a frequency of 10−8 mutations/bp [28]. The result of SHM in variable regions is increased affinity of the antibody for antigen, and the result of SHM in the switch region is increased double-strand breaks for CSR from IgM to IgG, IgA, and IgE. The absence of AID in humans leads to type II hyper-IgM syndrome [29], where individuals are at increased risk of disease because they can only produce low affinity antibodies of the IgM isotype. Contrary to intuition, deficiencies in MMR proteins actually lead to decreased mutagenesis in variable and switch regions [30, 31]. This occurs because SHM relies on a hijacked version of the MMR pathway to create mutations at A and T residues [32] (Figure 1).
Fig. 1. MMR proteins create mutations at A:T bp during SHM.
AID deaminates cytosine to uracil in immunoglobulin variable region DNA and generates a U:G mismatch that is recognized by the MSH2-MSH6 heterodimer. A single-strand DNA gap is produced at the mismatch by EXO1 and an unknown nick instigator. Monoubiquitinated PCNA homotrimer recruits the error-prone DNA pol η to fill in the gap, copying the original T with a G instead of an A. DNA replication results in the G:T mismatch being permanently affixed in one of the two daughter cells as a mutation to G:C.
The MutSα heterodimer binds to an AID-induced U:G mismatch and recruits a nick-creating nuclease. The identity of the nuclease is unknown, but it is unlikely to be the MutLα complex employed in canonical repair; possible suspects are examined in more detail in 2.2. This nuclease acts along with EXO1 to remove the mismatch and adjacent bases. Monoubiquitinated PCNA then encircles the gap and binds error-prone DNA pol η, which favors synthesis of mutations opposite A and T nucleotides, to fill in the gap. It is not known what cellular signals allow SHM to proceed in place of canonical MMR.
An analogous SHM process removes uracils in DNA via a distorted BER pathway [2]. UNG recognizes the uracil and employs its glycosylase activity to create an abasic site. APE creates a nick at the abasic site, which is filled in by Rev1 to create mutations at C and G bases. Ung−/− mice have a normal mutation frequency but decreased C:G transversions, while A:T mutagenesis is unchanged [33]. Other uracil glycosylases such as single-strand-selective monofunctional uracil-DNA glycosylase (SMUG1) [34, 35] and methyl-CpG binding domain 4 [36, 37] are not involved in generating abasic sites for SHM under normal physiological conditions.
Recent studies have also examined the roles of APE1 and APE2 endonucleases [38]. APE1 is highly expressed in resting or in vitro activated B cells, but is poorly expressed in germinal centers. APE1-haploinsufficient mice do not have an appreciably altered SHM frequency or spectrum. In contrast, APE2 is highly expressed in germinal centers, and APE2-deficient mice have a 50% decline in overall mutagenesis and a slight decline in mutations at A:T bases [38, 39]. Mice that are doubly-deficient for UNG and APE2 exhibit an additional 2-fold decline in A:T mutations. These unexpected results suggest that APE2 may have a role outside of the BER pathway, and could interact with additional glycosylases or MMR components. Alternatively, the doubly-deficient cells may have severely impaired cell cycle progression [40], which provides additional time to undergo faithful DNA repair.
2.2. Contribution of individual MMR components to SHM
Many details of the MMR-SHM pathway have been elucidated in knockout or mutant mouse models [32, 41] (Table 1). Msh2−/− [42, 43] and Msh6−/− [44, 45] mice have reduced mutation frequencies in variable regions relative to wild type mice, and their counterparts containing an inactivated ATPase domain feature a similar but less severe phenotype [46, 47]. The MutSα-deficient mice exhibit an altered mutation spectrum, with a 75-90% decrease in mutations at A:T bp [44, 45, 48]. A marked decline in A:T mutagenesis has also been observed in MSH6-deficient humans [49]. Exo1−/− mice have decreased SHM and mutations at A:T bp [50, 51]. An initial report indicated that an Exo1E109K/E109K knock-in mouse was catalytically inactive but underwent standard SHM, suggesting that separate exonuclease structural and functional roles exist [51]. However, recent studies have countered that EXO1-E109K is a fully functional protein in vitro, and that EXO1 enzymatic function is required for its interactions with MMR proteins [52, 53].-
Table 1.
MMR mutant mouse models in antibody diversification. Y, yes; N, no.
| MMR defect | Decreased SHM? | Decreased mutations at A:T? | Decreased CSR? | Notes | References |
|---|---|---|---|---|---|
| Msh2−/− | Y | Y | Y | [42, 43, 48, 101, 108] | |
| Msh2G674A | Y | Y | Y | inactive ATPase | [46] |
| Msh6−/− | Y | Y | Y | [44, 45, 110] | |
| Msh6T1217D | Y | Y | Y | inactive ATPase | [47] |
| Mlh1−/− | N | N | Y | [55, 101, 109] | |
| Mlh1G67R | N | N | Y | inactive ATPase | [111] |
| Pms2−/− | N | N | Y | [42, 56, 67, 101, 114] | |
| Pms2E702K | N | N | Y | inactive and unstable endonuclease | [66] |
| Msh3−/− | N | N | N | [44, 45, 110] | |
| Exo1−/− | Y | Y | Y | [50, 51] | |
| Exo1EK | N | N | N | conflicting enzymatic activity | [51][52, 53] |
| Ape1+/− | N | N | Y | [38, 98] | |
| Ape2y/m | Y | Y | Y? | conflicting CSR | [38, 39, 98] |
| PcnaK164R | Y | Y | Y? | no monoubiquitination, conflicting CSR | [60-62, 115] |
| Polh−/− | N | Y | N | [75-77, 94, 116] | |
| Polk−/− | N | N | N | backup for pol η | [93, 94, 117] |
| Ung−/− | N | N | Y | base excision repair | [33], [118] |
Mice that are doubly-deficient for MSH2 and MSH6 still exhibit the signature A:T mutation decline but have a large increase in C:G transitions, leading to an overall mutation frequency similar to wild type mice [54]. This may occur due to unrepaired uracils persisting in the DNA and being replicated directly across as C:G transitions. In contrast, deficiencies in MLH1 [55], MSH3 [44, 45], or PMS2 [42, 56] do not alter mutation spectrum, indicating that they are not essential for SHM via the MMR pathway. This stands in contrast to canonical MMR, where the MutL heterodimer is mandatory for DNA repair. Triple knockout Msh2−/−Msh3−/−Msh6−/− mice likewise do not show additional changes in mutation frequency or spectrum when compared to respective double knockouts [54].
Interactions of other MMR factors with the MutSα complex can have substantial effects on SHM. Post-translational modification of the PCNA sliding clamp is of special interest because it helps regulate the choice between error-free repair and error-prone SHM. PCNA that is polyubiquitinated at lysine 63 elicits a high-fidelity repair pathway [57, 58], while monoubiquitination at lysine 164 triggers SHM [59, 60]. Although the Pcna−/− genotype is embryonic lethal in mice, a PcnaK164R model was generated that cannot undergo monoubiquitination, and it exhibited a 90% decrease in A:T mutations [61, 62]. The remaining A:T mutations may be introduced through the activity of UNG and/or pol ζ, as discussed later in this manuscript. PCNA deubiquitination by USP1 [63, 64] also likely regulates SHM, although its role has not been extensively analyzed.
A major question that still remains concerning the MMR pathway in SHM is the identity of the nuclease responsible for providing the nick required prior to EXO1 activity. Nucleases that have been hypothesized to cause nick formation include PMS2 and APE. The MutLα complex may seem like a logical choice, as it interacts with MutSα during canonical MMR and its PMS2 subunit contains latent endonuclease activity [65, 66]. However, Pms2−/− mice have a normal SHM frequency [42, 56, 67], indicating that either PMS2 is not the major nuclease or that compensatory nucleases can act efficiently in its absence. Neuberger and Rada proposed that the glycosylase component of SMUG1 could serve as the nick instigator by creating an abasic site that is then cleaved by APE [34, 68]. Although SHM is unaltered in SMUG1-deficient mice, a small but consistent decrease in A:T mutations occurs in an Ung−/−Smug1−/− background. Additional studies will be necessary to identify the endonuclease involved in making nicks for MMR-SHM.
2.3. Generation of A:T mutations by pol η
Pol η is responsible for the majority of mutations at A:T bp during SHM [69, 70]. Pol η is a γ family translesion polymerase encoded by the Polh gene. It inserts nucleotides opposite adducts, including UV-caused cyclobutane pyrimidine dimers and cisplatin-generated crosslinks, via short-patch synthesis [71]. However, during SHM, pol η exhibits promiscuous fidelity when copying A and T bases on undamaged DNA [72]. Pol η has been shown to be catalytically activated upon binding to the MutSα heterodimer in vitro [73]. Pol η prefers to insert G opposite T on the transcribed strand, leading to A bases being mutated twice as frequently as T in variable and switch regions [74]. Polh−/− mice display an 85% reduction in A:T mutations, although the overall SHM frequency remains constant due to overcompensation by C:G mutations generated in the UNG pathway [75]. A similar alteration in mutation spectrum occurs in humans diagnosed with xeroderma pigmentosum variant syndrome, which is caused by defective pol η and results in a 4-fold decrease in A:T mutations [74, 76-78].
Although both Polh−/− and Msh2−/− or Msh6−/− mice display reduced A:T mutations, a distinct difference occurs in their respective mutation locations. Msh2−/− or Msh6−/− mice exhibit mutations targeted to AID-favored WGCW sequence hotspots, while Polh−/− mice have mutations distributed across the entire variable region [24, 69, 72, 75, 79]. MutSα is responsible for the recruitment of downstream proteins that introduce a single strand gap at the mutation site; in the absence of either of its components, mutations are targeted directly at the uracil site. Meanwhile, a deficiency of pol η does not interfere with the formation of a multi-base gap, allowing for mutations to be spread across a wider region and not focused on sequence hotspots.
The background A:T mutations present in Polh−/− individuals must be attributable to an additional promiscuous polymerase(s). Deficiencies in the non-replicative pols ι [80, 81], β [82, 83], λ [83, 84], μ [84], or θ [85] result in unchanged SHM. Pol ζ tends to insert tandem mutations [86, 87], while Rev1 undertakes the majority of G:C to C:G transversions via a distorted BER pathway [88-90]. The polymerase that creates G:C to T:A transversions is currently undefined. The remaining A:T mutations in Polh−/− mice typically consist of 50% T to G and A to C transversions, which matches the mutational spectrum of pol κ [69, 91, 92], although Polk−/− mice exhibit a normal SHM frequency and spectrum [93].
The polymerase responsible for generating the residual A:T mutations may be masked in the presence of pol η, so doubly-deficient mice were bred. Polh−/−Polk−/− mice showed a 93% reduction in A:T mutations relative to wild type, indicating that pol κ could contribute to mutagenesis in the absence of η [94]. It remains unclear what other polymerase(s) is responsible for the lingering A:T mutations in Polh−/− Polk−/− mice, although recent work suggests pol ζ may be involved. Msh2−/− and Msh6−/− mice both exhibit a decrease in ζ-induced tandem mutations, whereas the frequency of contiguous tandem mutations was unaffected in Ung−/− mice [86]. Pol ζ does not have a preferred mutation pattern and may be able to insert mutations at A:T in a Polh−/−Polk−/− environment.
Interestingly, Reynaud and colleagues showed that Msh2−/−Polh−/− mice have completely abolished A:T mutations [94]. This strongly suggests that the residual mutations present in the Msh2−/− or Msh6−/− mice are created through the activity of pol η in the UNG pathway, perhaps through a long-patch BER mechanism. This hypothesis is further supported by the mutational spectrum of Ung−/−Msh2−/− or Ung−/−Msh6−/− mice, which likewise lack mutations at A:T bases [24, 95, 96]. In summary, Pol η is clearly required for A:T mutagenesis in a normal physiological context.
3. MutSα and MutLα complexes assist in formation of switch junctions during CSR
An antibody's heavy chain constant region is directly involved in regulating its trafficking and binding to cellular receptors. Eight different isotypes are encoded in the murine Igh locus. Germline antibodies are exclusively IgM or IgD, but other isotypes are expressed through CSR in activated B cells [97]. In CSR, two switch region double-strand breaks are recombined, and this results in a change in antibody isotype, e.g., from IgM to IgG1. Switch regions are 3-9 kb long, and contain abundant WGCW hotspots alongside clusters of C bases on the transcribed strand, which allow for the formation of stable RNA-DNA hybrid structures during transcription. Thus, the DNA sequence of the switch regions promotes single-strand DNA for AID to bind the multiple hotspots and initiate deamination. This generates a profusion of uracils, which can be processed to create double-strand breaks for CSR.
BER proteins produce the majority of double-strand breaks. Ung−/− mice have a 95% reduction in CSR [33], while APE deficiency has a less prominent effect [39, 98]. Nearby BER-induced nicks on opposite strands simulate a double-strand break, which catalyzes recombination with double-strand breaks in other switch regions for CSR [99].
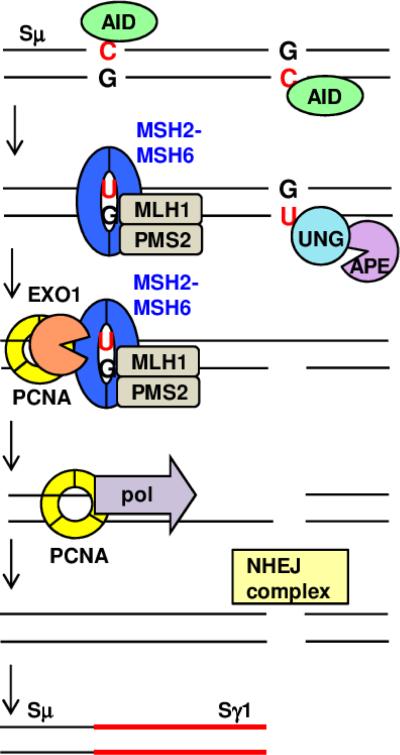
Alternatively, uracils may be spaced too far apart to permit spontaneous double-strand break formation, and instead require the assistance of MMR proteins (Figure 2). This idea is supported by data demonstrating significantly reduced double-strand breaks in the switch regions of MMR-deficient B cells [100]. Additionally, the isotype most severely affected by the deletion of MMR proteins is IgG2a, which contains less AID hotspots than the other isotypes [101].
Fig. 2. MMR proteins assist in the conversion of BER-induced single-strand nicks to double-strand break substrates for CSR.
AID-induced uracils in immunoglobulin switch region DNA can be bound by either BER or MMR proteins. In BER, UNG and APE remove the uracil and create a single-strand nick. MMR proteins MSH2-MSH6 and MLH1-PMS2 encounter a uracil on the opposite strand and attract EXO1 and PCNA to an adjacent nick. EXO1 excises the sequence between the nicks, leading to a 5’ overhang that is processed by PCNA and a translesion polymerase to create a blunt double-strand break. Alternatively, the overhang can be deleted by EXO1 or a 5’ flap endonuclease. An analogous process can occur with MMR proteins acting on a uracil located 3’ to the BER-induced nick (not shown). When double-strand breaks occur simultaneously in different switch regions, recombination produces a new switch junction mediated by NHEJ factors, as illustrated here between Sμ and Sγ1.
Whenever MMR and BER proteins are both involved in CSR [99, 102], one AID-induced uracil is processed conventionally by UNG and APE, resulting in a single-strand nick. A second, non-proximal uracil on the opposite strand is recognized by MutSα and MutLα. These proteins recruit EXO1 and PCNA, which bind to a nick generated by an undefined nuclease. EXO1 will excise one strand of DNA between the two nicks, and can act in either the 5’ → 3’ or 3’ → 5’ direction depending on the relative position of the nicks. The resulting overhang can be filled in by a PCNA-translesion polymerase complex or removed by a flap endonuclease or EXO1. This creates double-strand break substrates for CSR.
These double-strand DNA breaks are usually repaired through non-homologous end joining (NHEJ), which uses either blunt ends or ends with short microhomology [103]. Alternative-end joining featuring longer microhomologies at the site of joining can also occur, but is commonly associated with nonproductive intra-switch recombination [104, 105]. The cell cycle phase also regulates break resolution by CSR. NHEJ is active throughout the cell cycle, while alternative-end joining favors late S or G2 phase [106, 107]. Since AID-induced double-strand breaks occur mainly during the G1 phase [100], break resolution generally proceeds through NHEJ.
A role for many MMR factors in CSR has been elucidated by examining changes in knockout mouse models (Table 1). Mice that are deficient for MSH2 [43, 101, 108, 109], MSH6 [45, 110], MLH1 [101, 109], or PMS2 [56, 101] all exhibit a 50% or greater reduction in switching in spleen cells stimulated ex vivo, or in serum in vivo. The ATPase domain of MSH2 [46], MSH6 [47], or MLH1 [111] is likewise utilized; mice with inactivated ATPase sites experienced decreased CSR. The MSH2 and MLH1 ATPase mutant mice possessed additional changes in microhomologies, suggesting that their ATPase activity is a prerequisite for efficient CSR. MSH3 has also been examined; deletion of the protein does not alter CSR, similar to its noninvolvement in SHM [45, 110].
Many other factors that interact with the MMR proteins have been tested for their involvement in CSR. Exo1−/− mice show a 70% decline in CSR and altered microhomology similar to Msh2−/− [50], implying that both proteins are involved in break formation. Monoubiquitinated PCNA may play a role, but conflicting results have been reported in independently derived PcnaK164R mouse models [60, 61]. Although pol η is required for the MMR pathway during SHM, it is not involved in CSR [75]. Future studies will help to extract more information concerning what other cellular interactions oversee CSR.
4. Conclusion and future directions
SHM and CSR are unique in that they commandeer seemingly faithful repair pathways and instead use them to create mutations and strand breaks. Much work remains to be done to determine how B cells choose to respond to AID-induced uracils. Recently published studies suggest that AID is targeted to the Ig locus through its interactions with a combination of transcription complexes and specific sequence elements [112, 113]. Once the uracil is formed, cells must decide whether to undergo repair or mutagenesis using either BER or MMR components. It is unclear whether MutSα and UNG work together or compete to manage uracils. Other poorly understood issues include why mutagenesis occurs in both variable and switch regions, while double-strand breaks are exclusive to switch regions, and why certain proteins participate in one process but not the other.
Highlights.
Immunoglobulin diversification utilizes mismatch repair proteins at U:G mismatches.
MutSα and DNA polymerase η generate mutations during somatic hypermutation.
MutSα and MutLα regulate class switch recombination.
Acknowledgements
We thank Robert Maul for helpful discussions and feedback. This research was supported entirely by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Abbreviations
- AID
activation-induced deaminase
- APE
apurinic/apyrimidinic endonuclease
- BER
base excision repair
- bp
base pair
- CSR
class switch recombination
- D
diversity gene segment
- EXO1
exonuclease 1
- Ig
immunoglobulin
- J
joining gene segment
- pol
polymerase
- MMR
mismatch repair
- NHEJ
nonhomologous end joining
- SHM
somatic hypermutation
- SMUG1
single-strand-selective monofunctional uracil-DNA glycosylase
- UNG
uracil DNA glycosylase
- V
variable gene segment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Subrahmanyam R, Sen R. RAGs' eye view of the immunoglobulin heavy chain gene locus. Semin Immunol. 2010;22:337–345. doi: 10.1016/j.smim.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 6.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 7.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, Reynaud CA. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 8.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han S, Zheng B, Takahashi Y, Kelsoe G. Distinctive characteristics of germinal center B cells. Semin Immunol. 1997;9:255–260. doi: 10.1006/smim.1997.0081. [DOI] [PubMed] [Google Scholar]
- 10.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 11.Michael N, Martin TE, Nicolae D, Kim N, Padjen K, Zhan P, Nguyen H, Pinkert C, Storb U. Effects of sequence and structure on the hypermutability of immunoglobulin genes. Immunity. 2002;16:123–134. doi: 10.1016/s1074-7613(02)00261-3. [DOI] [PubMed] [Google Scholar]
- 12.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohli RM, Maul RW, Guminski AF, McClure RL, Gajula KS, Saribasak H, McMahon MA, Siliciano RF, Gearhart PJ, Stivers JT. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J Biol Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajula KS, Huwe PJ, Mo CY, Crawford DJ, Stivers JT, Radhakrishnan R, Kohli RM. High-throughput mutagenesis reveals functional determinants for DNA targeting by activation-induced deaminase. Nucleic Acids Res. 2014;42:9964–9975. doi: 10.1093/nar/gku689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canugovi C, Samaranayake M, Bhagwat AS. Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J. 2009;23:34–44. doi: 10.1096/fj.08-115352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Fan M, Kalis S, Wei L, Scharff MD. A source of the single-stranded DNA substrate for activation-induced deaminase during somatic hypermutation. Nature communications. 2014;5:4137. doi: 10.1038/ncomms5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, Papavasiliou FN. Switch recombination and somatic hypermutation are controlled by the heavy chain 3' enhancer region. J Exp Med. 2009;206:2613–2623. doi: 10.1084/jem.20091280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka A, Shen HM, Ratnam S, Kodgire P, Storb U. Attracting AID to targets of somatic hypermutation. J Exp Med. 2010 doi: 10.1084/jem.20090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramiro A, San-Martin BR, McBride K, Jankovic M, Barreto V, Nussenzweig A, Nussenzweig MC. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- 20.Crouse GF. Non-canonical actions of MMR. DNA repair. doi: 10.1016/j.dnarep.2015.11.020. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiricny J. The multifaceted mismatch-repair system, Nature reviews. Molecular cell biology. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 22.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chahwan R, Edelmann W, Scharff MD, Roa S. Mismatch-mediated error prone repair at the immunoglobulin genes. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2011;65:529–536. doi: 10.1016/j.biopha.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Maul RW, Gearhart PJ. Controlling somatic hypermutation in immunoglobulin variable and switch regions. Immunol Res. 2010;47:113–122. doi: 10.1007/s12026-009-8142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5' boundary is near the promoter, and 3' boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogne M, Denizot Y. The IgH 3' regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 29.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 30.Saribasak H, Gearhart PJ. Does DNA repair occur during somatic hypermutation? Semin Immunol. 2012;24:287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chahwan R, Edelmann W, Scharff MD, Roa S. AIDing antibody diversity by error-prone mismatch repair. Semin Immunol. 2012;24:293–300. doi: 10.1016/j.smim.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saribasak H, Rajagopal D, Maul RW, Gearhart PJ. Hijacked DNA repair proteins and unchained DNA polymerases. Philos Trans R Soc Lond B Biol Sci. 2009;364:605–611. doi: 10.1098/rstb.2008.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 34.Dingler FA, Kemmerich K, Neuberger MS, Rada C. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts A:T substitutions during somatic mutation. Eur J Immunol. 2014 doi: 10.1002/eji.201444482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doseth B, Ekre C, Slupphaug G, Krokan HE, Kavli B. Strikingly different properties of uracil-DNA glycosylases UNG2 and SMUG1 may explain divergent roles in processing of genomic uracil. DNA repair. 2012;11:587–593. doi: 10.1016/j.dnarep.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Bardwell PD, Martin A, Wong E, Ziqiang L, Edelmann W, Scharff MD. Cutting Edge: The G-U mismatch glycosylase methyl-CpG binding domain 4 is dispensable for somatic hypermutation and class switch recombination. J Immunol. 2003;170:1620–1624. doi: 10.4049/jimmunol.170.4.1620. [DOI] [PubMed] [Google Scholar]
- 37.Grigera F, Bellacosa A, Kenter AL. Complex relationship between mismatch repair proteins and MBD4 during immunoglobulin class switch recombination. PLoS One. 2013;8:e78370. doi: 10.1371/journal.pone.0078370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stavnezer J, Linehan EK, Thompson MR, Habboub G, Ucher AJ, Kadungure T, Tsuchimoto D, Nakabeppu Y, Schrader CE. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci U S A. 2014;111:9217–9222. doi: 10.1073/pnas.1405590111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabouri Z, Okazaki IM, Shinkura R, Begum N, Nagaoka H, Tsuchimoto D, Nakabeppu Y, Honjo T. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol. 2009;21:947–955. doi: 10.1093/intimm/dxp061. [DOI] [PubMed] [Google Scholar]
- 40.Guikema JE, Gerstein RM, Linehan E, Cloherty E, Evan-Browning E, Tsuchimoto D, Nakabeppu Y, Schrader CE. Apurinic/apyridimic endonuclease 2 is necessary for normal B cell development and recovery of lymphoid progenitors after chemotherapeutic challenge. J Immunol. 2011;186:1943–1950. doi: 10.4049/jimmunol.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winter DB, Gearhart PJ. Altered spectra of hypermutation in DNA repair-deficient mice. Philos Trans R Soc Lond B Biol Sci. 2001;356:5–11. doi: 10.1098/rstb.2000.0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey S, Bertocci B, Delbos F, Quint L, Weill JC, Reynaud CA. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9:127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 43.Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- 44.Wiesendanger M, Kneitz B, Edelmann W, Scharff MD. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med. 2000;191:579–584. doi: 10.1084/jem.191.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin A, Li Z, Lin DP, Bardwell PD, Iglesias-Ussel MD, Edelmann W, Scharff MD. Msh2 ATPase activity is essential for somatic hypermutation at A-T basepairs and for efficient class switch recombination. J Exp Med. 2003;198:1171–1178. doi: 10.1084/jem.20030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Zhao C, Iglesias-Ussel MD, Polonskaya Z, Zhaung M, Yang G, Luo Z, Edelmann W, Scharff MD. The mismatch repair protien Msh6 influences the in vivo AID targeting to the Ig locus. Immunity. 2006;24:393–403. doi: 10.1016/j.immuni.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Phung QH, Winter DB, Cranston A, Tarone RE, Bohr VA, Fishel R, Gearhart PJ. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J Exp Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gardes P, Forveille M, Alyanakian M-A, Aucouturier P, Ilencikova D, Leroux D, Rahner N, Mazerolles A, Kracker S, Durandy A. Human MSH6 deficiency is associated with impaired antibody maturation. J Immunol. 2012;188:2023–2029. doi: 10.4049/jimmunol.1102984. [DOI] [PubMed] [Google Scholar]
- 50.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, Scharff MD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 51.Schaetzlein S, Chahwan R, Avdievich E, Roa S, Wei K, Eoff RL, Sellers RS, Clark AB, Kunkel TA, Scharff MD, Edelmann W. Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc Natl Acad Sci U S A. 2013;110:E2470–E2479. doi: 10.1073/pnas.1308512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bregenhorn S, Jiricny J. Biochemical characterization of a cancer-associated E109K missense variant of human exonuclease 1. Nucleic Acids Res. 2014;42:7096–7103. doi: 10.1093/nar/gku419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shao H, Baitinger C, Soderblom EJ, Burdett V, Modrich P. Hydrolytic function of Exo1 in mammalian mismatch repair. Nucleic Acids Res. 2014;42:7104–7112. doi: 10.1093/nar/gku420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roa S, Li Z, Peled JU, Zhao C, Edelmann W, Scharff MD. MSH2/MSH6 complex promotes error-free repair of AID-induced dU:G mispairs as well as error-prone hypermutation of A:T sites. PLoS One. 2010;5:e11182. doi: 10.1371/journal.pone.0011182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phung QH, Winter DB, Alrefai R, Gearhart PJ. Hypermutation in Ig V genes from mice deficient in the MLH1 mismatch repair protein. J Immunol. 1999;162:3121–3124. [PubMed] [Google Scholar]
- 56.Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology-mediated mechanism of Ig class switch recombination. Proc Natl Acad Sci USA. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 58.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiqutin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 59.Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roa S, Avdievich E, Peled JU, Maccarthy T, Werling U, Kuang FL, Kan R, Zhao C, Bergman A, Cohen PE, Edelmann W, Scharff MD. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc Natl Acad Sci USA. 2008;105:16248–16253. doi: 10.1073/pnas.0808182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langerak P, Nygren AO, Krijger PH, van den Berk PC, Jacobs H. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J Exp Med. 2007;204:1989–1998. doi: 10.1084/jem.20070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krijger PH, Langerak P, van den Berk PC, Jacobs H. Dependence of nucleotide substitutions on Ung2, Msh2, and PCNA-Ub during somatic hypermutation. J Exp Med. 2009;206:2603–2611. doi: 10.1084/jem.20091707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang T, Nijman S, Mirchandani K, Galardy P, Cohn M, Haas W, Gygi S, Ploegh H, Bernards R, D'Andrea A. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nature cell biology. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 64.Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, Reissner T, Lee K, Geacintov NE, Carell T, Myung K, Tateishi S, D'Andrea A, Jacobs H, Livneh Z. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kadyrov F, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 66.van Oers J, Roa S, Werling U, Liu Y, Genschel J, Hou H, Sellers R, Modrich P, Scharff MD, Edelmann W. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci U S A. 2010;107:13384–13389. doi: 10.1073/pnas.1008589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winter DB, Phung QH, Umar A, Baker SM, Tarone RE, Tanaka K, Liskay RM, Kunkel TA, Bohr VA, Gearhart PJ. Altered spectra of hypermutation in antibodies from mice deficient for the DNA mismatch repair protein PMS2. Proc Natl Acad Sci USA. 1998;95:6953–6958. doi: 10.1073/pnas.95.12.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maul RW, Gearhart PJ. Refining the Neuberger model: Uracil processing by activated B cells. Eur J Immunol. 2014;44:1913–1916. doi: 10.1002/eji.201444813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J Exp Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 72.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 73.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J Immunol. 2005;174:7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 75.Martomo SA, Yang WW, Wersto RP, Ohkumo T, Kondo Y, Yokoi M, Masutani C, Hanaoka F, Gearhart PJ. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc Natl Acad Sci USA. 2005;102:8656–8661. doi: 10.1073/pnas.0501852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 77.Zeng X, Negrete GA, Kasmer C, Yang WW, Gearhart PJ. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J Exp Med. 2004;199:917–924. doi: 10.1084/jem.20032022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Faili A, Aoufouchi S, Weller S, Vuillier F, Stary A, Sarasin A, Reynaud CA, Weill JC. DNA polymerase eta is involved in hypermutation occurring during immunoglobulin class switch recombination. J Exp Med. 2004;199:265–270. doi: 10.1084/jem.20031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reynaud CA, Delbos F, Faili A, Gueranger Q, Aoufouchi S, Weill JC. Competitive repair pathways in immunoglobulin gene hypermutation. Philos Trans R Soc London B Biol Sci. 2009;364:613–619. doi: 10.1098/rstb.2008.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J Exp Med. 2003;198:635–643. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martomo SA, Yang WW, Vaisman A, Maas A, Yokoi M, Hoeijmakers JH, Hanaoka F, Woodgate R, Gearhart PJ. Normal hypermutation in antibody genes from congenic mice defective for DNA polymerase iota. DNA Repair (Amst) 2006;5:392–398. doi: 10.1016/j.dnarep.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 82.Esposito G, Texido G, Betz UA, Gu H, Muller W, Klein U, Rajewsky K. Mice reconstituted with DNA polymerase beta-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc Natl Acad Sci U S A. 2000;97:1166–1171. doi: 10.1073/pnas.97.3.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schrader CE, Linehan EK, Ucher AJ, Bertocci B, Stavnezer J. DNA polymerases beta and lambda do not directly affect Ig variable region somatic hypermutation although their absence reduces the frequency of mutations. DNA Repair (Amst) 2013;12:1087–1093. doi: 10.1016/j.dnarep.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bertocci B, De Smet A, Flatter E, Dahan A, Bories JC, Landreau C, Weill JC, Reynaud CA. Cutting edge: DNA polymerases mu and lambda are dispensable for Ig gene hypermutation. J Immunol. 2002;168:3702–3706. doi: 10.4049/jimmunol.168.8.3702. [DOI] [PubMed] [Google Scholar]
- 85.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–1608. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saribasak H, Maul RW, Cao Z, Yang WW, Schenten D, Kracker S, Gearhart PJ. DNA polymerase zeta generates tandem mutations in immunoglobulin variable regions. J Exp Med. 2012;209:1075–1081. doi: 10.1084/jem.20112234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daly J, Bebenek K, Watt DL, Richter K, Jiang C, Zhao ML, Ray M, McGregor WG, Kunkel TA, Diaz M. Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase zeta activity. J Immunol. 2012;188:5528–5537. doi: 10.4049/jimmunol.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, de Wind N. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med. 2006;203:319–323. doi: 10.1084/jem.20052227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang J-Y. A critical role for REV1 in regulating the induciton of C:G transitions and A:T mutations during Ig gene hypermutation. J Immunol. 2009;183:1846–1850. doi: 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- 90.Krijger PH, Tsaalbi-Shtylik A, Wit N, Van den Berk P, De Wind N, Jacobs H. Rev1 is essential in generating C to G transversions downstream of the UNG2 pathway but not the Msh2+Ung2 hybrid pathway. Eur J Immunol. 2013;43:2765–2770. doi: 10.1002/eji.201243191. [DOI] [PubMed] [Google Scholar]
- 91.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y, Yuan F, Wu X, Wang M, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schenten D, Gerlach VL, Guo C, Velasco-Miguel S, Hladik CL, White CL, Friedberg EC, Rajewsky K, Esposito G. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur J Immunol. 2002;32:3152–3160. doi: 10.1002/1521-4141(200211)32:11<3152::AID-IMMU3152>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 94.Faili A, Stary A, Delbos F, Weller S, Aoufouchi S, Sarasin A, Weill JC, Reynaud CA. A backup role of DNA polymerase kappa in Ig gene hypermutation only takes place in the complete absence of DNA polymerase eta. J Immunol. 2009;182:6353–6359. doi: 10.4049/jimmunol.0900177. [DOI] [PubMed] [Google Scholar]
- 95.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J Immunol. 2006;177:5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- 97.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 98.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 101.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J Exp Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stavnezer J, Schrader CE. Mismatch repair converts AID-instigated nicks to double-strand breaks for antibody class-switch recombination. TRENDS in Genetics. 2006;22:23–28. doi: 10.1016/j.tig.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 103.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 104.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 106.Wu W, Wang M, Wu W, Singh SK, Mussfeldt T, Iliakis G. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA repair. 2008;7:329–338. doi: 10.1016/j.dnarep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 107.Singh SK, Wu W, Zhang L, Klammer H, Wang M, Iliakis G. Widespread dependence of backup NHEJ on growth state: ramifications for the use of DNA-PK inhibitors. Int J Radiat Oncol Biol Phys. 2011;79:540–548. doi: 10.1016/j.ijrobp.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 108.Ehrenstein MR, Neuberger MS. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schrader CE, Vardo J, Stavnezer J. Mlh1 can function in antibody class switch recombination independently of Msh2. J Exp Med. 2003;197:1377–1383. doi: 10.1084/jem.20022190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelmann W, Scharff MD. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J Exp Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chahwan R, van Oers J, Avdievich E, Zhao C, Edelmann W, Scharff MD, Roa S. The ATPase activity of MLH1 is required to orchestrate DNA double-strand breaks and end processing during class switch recombination. J Exp Med. 2012;209:671–678. doi: 10.1084/jem.20111531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maul RW, Cao Z, Venkataraman L, Giorgetti CA, Press JL, Denizot Y, Du H, Sen R, Gearhart PJ. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo-activated cells. J Exp Med. 2014;211:2297–2306. doi: 10.1084/jem.20131512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Q, Oliveira TY, Jankovic M, Silva I, Hakim O, Yao K, Gazumyan A, Mayer C, Pavri R, Casellas R, Nussenzweig M, Robbiani DF. Epigenetic targeting of activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2014;111:18667–18672. doi: 10.1073/pnas.1420575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kong Q, Maizels N. PMS2-deficiency diminishes hypermutation of a lambda1 transgene in young but not older mice. Mol Immunol. 1999;36:83–91. doi: 10.1016/s0161-5890(99)00027-9. [DOI] [PubMed] [Google Scholar]
- 115.Krijger P, van den Berk P, Wit N, Langerak P, Jansen J, Reynaud C-A, de Wind N, Jacobs H. PCNA ubiquitination-independent activation of polymerase η during somatic hypermutation and DNA damage tolerance. DNA repair. 2011;10:1051–1059. doi: 10.1016/j.dnarep.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 116.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, J OW. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J Biol Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 117.Shimizu T, Shinkai Y, Ogi T, Ohmori H, Azuma T. The absence of DNA polymerase kappa does not affect somatic hypermutation of the mouse immunoglobulin heavy chain gene. Immunol Lett. 2003;86:265–270. doi: 10.1016/s0165-2478(03)00046-4. [DOI] [PubMed] [Google Scholar]
- 118.Zahn A, Daugan M, Safavi S, Godin D, Cheong C, Lamarre A, Di Noia J. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J Immunol. 2013;190:5949–5960. doi: 10.4049/jimmunol.1202711. [DOI] [PubMed] [Google Scholar]