Abstract
We have currently entered a genomic era of cancer research which may soon lead to a genomic era of cancer treatment. Patient DNA sequencing information may lead to a personalized approach to managing an individual’s cancer as well as future cancer risk. The success of this approach, however, begins not necessarily in the clinician’s office, but rather at the laboratory bench of the basic scientist. The basic scientist plays a critical role since the DNA sequencing information is of limited use unless one knows the function of the gene that is altered and the manner by which a sequence alteration affects that function. The role of basic science research in aiding the clinical management of a disease is perhaps best exemplified by considering the case of Lynch syndrome, a hereditary disease that predisposes patients to colorectal and other cancers. This review will examine how the diagnosis, treatment and even prevention of Lynch syndrome-associated cancers has benefitted from extensive basic science research on the DNA mismatch repair genes whose alteration underlies this condition.
Keywords: Lynch syndrome, mismatch repair, colorectal cancer, personalized medicine, microsatellite instability, chemotherapy
1. Introduction
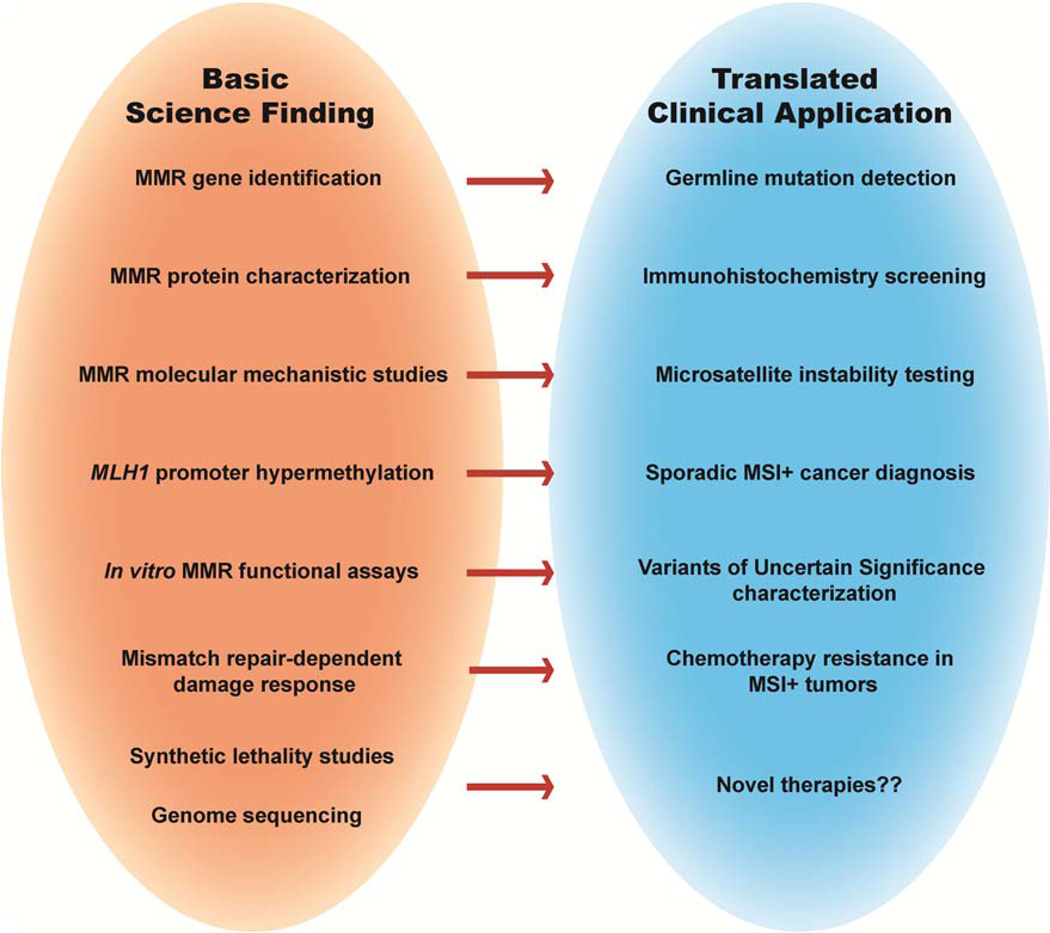
Lynch syndrome (LS) is the most prevalent hereditary colorectal cancer (CRC)-predisposition syndrome resulting in approximately 30,000 cases of CRC per year (1). This syndrome has also been referred to as hereditary non-polyposis colon cancer (HNPCC), however, the recognition that numerous extracolonic cancers can develop in these patients, such as endometrial, ovarian, stomach, pancreatic and multiple other cancers, has led many to discontinue using this nomenclature (2). Successful management of LS patients begins with a proper diagnosis of the condition, and includes often times aggressive cancer prevention approaches and appropriate decisions about cancer treatment. Understanding the molecular changes underlying tumor etiology aides the clinical decision-making process at each step. The discovery that LS is caused by inherited mutations in genes of the DNA mismatch repair (MMR) pathway has been tremendously important for the management of this disease. Basic science research on the MMR pathway predated its link to human cancer and only accelerated after the connection was established. The information learned in the laboratory has had implications for LS diagnosis, prevention and treatment. Whereas many of the minireviews in this special issue will deal with specific aspects of MMR molecular mechanism, this review will highlight examples of how this mechanistic information has been put to use in the clinic (Fig. 1). In addition, minireviews by Peña-Diaz & Rasmussen; Li, Pearlman & Hsieh; Sijmons & Hofstra; and Begum & Martin will further elaborate on some of the connections between basic science research and clinical advances highlighted here.
Fig. 1.
Some of the major basic science research breakthroughs in the DNA mismatch repair field and the clinical application that resulted directly or indirectly.
2. History of Lynch syndrome
The first patient that we now recognize as likely having LS was a German immigrant who settled in Michigan in the mid 1800’s (3). Before dying of cancer at the age of 60, the man fathered 10 children, 6 of whom also died of cancer. One of his descendants was a young seamstress who reported to her employer, Aldred Warthin, then Chairman of the Department of Pathology at the University of Michigan, that she was very worried that she might die of cancer since so many members of her family had previously died of cancer. This conversation led Warthin to collect information on her family history and create a pedigree describing the multiple cancers that affected successive generations in her family. Warthin further examined the cancer cases that came through his pathology laboratory and noted that around 15% of those cases had some family history of cancer (4). He concluded that there must be a hereditary effect on some cancers.
In 1962, Henry Lynch, an internal medicine resident in Nebraska with an interest in genetics, similarly met a patient recovering from delirium tremens who blamed his heavy drinking on fears of dying from colorectal cancer due to the many family members who died of cancer (2). As did Warthin, Lynch proceeded to develop a family history of this patient, which showed multiple cases of CRC which lacked the distinct polyposis phenotype of familial adenomatous polyposis (FAP), a known CRC-predisposition syndrome at that time. Soon after, Lynch was made aware of the work at the University of Michigan describing similar families and began to argue for the recognition of a new syndrome of clustered cancers in families, referred to at the time as cancer family syndrome (CFS). That this syndrome had a genetic basis was a contentious issue for many years as the accepted dogma was that environmental factors, commonly shared within families, was the main cause of cancer.
3. Into The Molecular Era of LS Diagnosis
3.1 The Molecular Genetics of LS
As Lynch and other researchers accumulated information data on CFS families around the world accumulated over the next 30 years, acknowledgement of this condition, eventually to be referred to as LS, became more common. The diagnosis of LS depended on knowledge of family history to determine the extent of cancer clustering in the family (see the minireview of Sijmons and Hofstra in this issue). The presence of a family history of cancer and/or an early age of cancer diagnosis remains the first clue that leads to a clinical suspicion of LS. The average LS patient develops cancer by the age of 45, two decades earlier than in the general population. To aid the diagnosis of LS, an International Collaborative Group on HNPCC devised clinical criteria known as the Amsterdam Criteria (5) that established rules based on age of diagnosis and number of cancers across multiple generations in a family. Those criteria were expanded in 1999 to account for the extracolonic cancers frequently observed in LS families (6). However, a leap forward in LS diagnosis emerged from the discovery in 1993 that almost all LS tumors displayed a form of nucleotide-level genomic instability called microsatellite instability (MSI) (7–9). While using polymorphic microsatellite markers to identify regions of loss of heterozygosity, it was observed that LS-associated tumors contained additional expansions or contractions in these repeat sequences. The significance of these findings was not immediately obvious to clinicians and even cancer researchers. However, some basic scientists did recognize this phenotype as one they had observed in lower organisms with defects in DNA repair and recombination pathways.
MSI, or the increased tendency of tandem repeat sequences to undergo small insertion or deletion loop (IDL) mutations, was first noted in the 1970s in bacteriophage (10) and was proposed to occur when denatured repeats reannealed out of register leading to a bulge of bases from the duplex DNA (11). This instability was enhanced when repeat-containing sequences were introduced into mutS or mutL Escherichia coli strains defective for MMR, a pathway already recognized to be involved in repairing mispaired bases (12). Similarly, studies in Saccharomyces cerevisiae showed that mutations in MSH2, MLH1 or PMS1 resulted in a several hundred fold increase in tandem repeat instability, further linking the MMR pathway with MSI (13). Upon report of increased MSI in LS cancers, basic scientists studying MMR in lower organisms turned their attention to discovering a homologous pathway in human cells. One of the first reports by Parsons et al. (14), showed that cancer cell lines derived from LS patients failed to repair small IDLs as well as single basepair mismatches. This report suggested that LS was a disease of defective MMR. During the same time period, Fishel et al., identified the human MutS homolog MSH2 and suggested that mutations in this gene were causative for LS (15) as did Leach et al., who identified deleterious mutations within the human MSH2 gene (16). The following year, two groups cloned the human MutL homolog MLH1 and found deleterious mutations within LS families (17,18). In addition to these two genes, another human MutS homolog MSH6 was later discovered and reported to be altered in the germline of LS patients (19). Finally, a small subset of LS patients was shown to have mutations in the MutL homolog PMS2 (20,21). Thus, basic science research on DNA repair in yeast and bacteria provided the crucial insights necessary to determine the genetic basis for LS, establishing it as a genetic disease and vindicating the work of Warthin, Lynch and others. Importantly, this discovery also sparked a new field of cancer research that elucidates the role of DNA repair and genome stability in preventing tumorigenesis.
In addition to establishing a genetic basis for LS, linking defective MMR with the disease led to the current molecular era of LS diagnosis. While the Amsterdam criteria assisted in identifying LS patients, their use depended on the patient having accurate knowledge of their family history, and even then may not capture all cases (22). However, the ability to now assess a tumor for molecular markers of defective MMR can provide a diagnostic clue. MSI detection, via a simple PCR screen, is commonly used as a first screen for defective MMR in suspected LS patients (23,24). In addition, as the majority of germline mutations in MMR genes in LS patients result in loss of stable protein, immunohistochemistry (IHC) analysis of the tumors is also performed using antibodies against the four major MMR proteins (25). Guidelines, similar to the Amsterdam criteria, though less stringent, were developed to identify those patients who should undergo these molecular tests (24). Termed the Bethesda Guidelines, they use personal history and clinical features to identify suspected LS patients. Today, given the relatively inexpensive nature of the MSI and IHC molecular screens, many health centers have moved to a universal screening approach, testing all new CRC patients for MMR defects to identify possible LS cases, regardless of age or family history (22,26–30).
3.2 MMR-defective sporadic colorectal cancer
The detection of MSI in a tumor does not necessarily mean the patient suffers from LS. Between 10–40% of sporadic colon, endometrial, ovarian and other cancers display MSI as well indicating that MMR can be disrupted in sporadic cancers. The loss of MMR function in sporadic tumors was shown in multiple laboratories to result primarily from hypermethylation of the MLH1 promoter, leading to its reduced expression (31–33). Thus, to distinguish LS patients from sporadic cancer patients, the next step in the diagnostic process is to sequence the four major MMR genes using genomic DNA usually from a blood sample of the affected individual. Detection of a deleterious, germline mutation in one of the four major MMR genes often serves as the ultimate confirmation of an LS diagnosis. Thus, the use of DNA sequencing for personalized medicine has already been occurring in the LS field for over a decade.
3.3 Variants of Uncertain Significance
A publically available database of all known cancer-causing mutations in LS curated by the International Society for Gastrointestinal and Hereditary Tumors shows that 42% of LS cases are caused by mutations in MLH1, 33% in MSH2, 18% in MSH6 and 7.5% in PMS2 (34). While a clearly deleterious mutation such as a deletion or frameshift can confirm an LS diagnosis, about 20–30% of the mutations detected are missense variants (35,36). The pathogenic significance of these missense variants cannot necessarily be assumed preventing the clinician or genetic counselor from making a definitive diagnosis. These missense variants, termed variants of uncertain significance (VUS), have been a source of intensive investigation in the MMR field (for more in depth discussion of this topic, please refer to Peña-Diaz & Rasmussen in this issue). Recently, a group of clinicians, genetic counselors and basic scientists came together to devise a classification scheme for assessing the clinical significance of MMR VUS (37). This classification scheme relies on an accumulation of clinical and genetic data about the patient such as whether the variant segregates with the affected family members in a suspected LS family and whether it associates with a tumor that displays features of defective MMR such as MSI. In addition to these clinical and genetic data, classification also depends on a functional assessment of the variant-containing MMR protein performed in a basic science laboratory. Due to the extensive basic science research performed over the last few decades, much is known about the molecular mechanism of the MMR pathway. This information has been useful for developing functional assays for testing VUS.
The most common assays have been in vitro biochemical assays. The ability to reconstitute the entire repair of a single basepair mismatch in the test tube using cell extracts (38) or recombinant proteins (39,40) has allowed researchers to isolate the effects of individual variants in the MMR process (41–45). In addition, assays that isolate individual steps in this pathway such as DNA mismatch binding, MMR protein-protein interactions or ATP processing have been used (35,45–48). While in vitro assays may detect specific defects caused by a VUS, normal activity in these assays does not necessarily mean the variant is neutral. Not all functional aspects by which a variant can cause a defect in MMR are captured in these assays. Cellular localization, chromatin interaction and other protein-protein interactions which may be important for MMR function in vivo are not assessed. Thus, testing the effects of VUS function in a cellular model may be necessary to examine variants with no phenotype in in vitro studies. Early attempts to study VUS in cells involved transient transfections in transformed human cell lines (49–51). These early studies demonstrated that certain variants had effects on MMR protein stability which could affect MMR function. Transfection of GFP-tagged MMR variants into NIH-3T3 primary cells was used to identify variants that affected the nuclear localization of the MMR protein complexes, providing another functional assay that cannot be assessed in the test tube (48,52,53). As the ability to carefully control expression levels of the variant-containing protein via transient transfection is limited, more recent studies have examined other means of introducing the variant transgenes such as the use of lentiviral vectors (54) or direct variant knock-in into the endogenous gene in mouse embryonic stem cells (55–57). The significance of the VUS problem in LS underscores the challenges that await the broader clinical community as the use of genome sequencing information for guiding diagnosis and clinical management increases. For there to still exist so much uncertainty for a disease like LS whose genetic basis is so well understood involving a pathway whose function is so well understood causes one to pause when contemplating the disease significance of a sequence variant in a gene that is not as well understood as the MMR genes. However, the strategies applied in classifying MMR VUS in LS may serve as a guide for meeting those challenges.
4. Mechanistic lessons applied to treatment of MMR-defective cancers
4.1 The MMR-dependent damage response
Information about the MMR pathway from the basic science laboratory has been essential for guiding the diagnosis of LS, however, the current challenge for many laboratories is learning information that will help guide treatment and even prevention of MMR-defective cancers. The observation that MMR-deficient cells are more resistant to the cytotoxic effects of certain DNA damaging agents used in conventional chemotherapy was a major discovery in this respect (58–61). The first evidence of a MMR-dependent damage response was demonstrated in MMR-deficient E. coli that were shown to be resistant to the cytotoxic effects of the DNA alkylating agent N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) (62). This finding was later confirmed in human cells (63) and has become the best characterized MMR-dependent damage response (for an in depth review, refer to Li, Pearlman & Hsieh in this issue). The major cytotoxic lesion created by MNNG is the O6-methylguanine (MeG) which is commonly mispaired with thymine during replication resulting in a potentially mutagenic MeG-T mispair. The MMR pathway recognizes these lesions and removes the mismatched thymine in the daughter strand. However, if the MeG lesion in the parental strand is not removed, usually by the direct-repair protein methylguanine methyltransferase (MGMT) (64), the polymerase will likely misincorporate a thymine again upon resynthesis. This has been proposed to lead to repeated futile cycles of MMR recognition and excision and the ultimate generation of an unreplicated gap opposite the MeG (65–67). When the cell enters the next cell cycle, a new replication fork will encounter this gap and convert it into a lethal double strand break. Thus, processing of a MeG-T mispair by the MMR pathway creates secondary DNA damage that ultimately causes cell death. In addition to the futile cycle model, researchers have demonstrated protein-protein interactions between the MMR proteins and key DNA damage signaling molecules such as ATR, ATM, CHK1 and CHK2 (68–72) suggesting a more direct role for the MMR proteins in triggering a damage response. It is proposed that the MMR proteins, following lesion recognition, can recruit damage signaling kinases to sites of damaged DNA. Both mechanisms indicate that MMR plays a protective role in removing damaged cells to reduce the risk of mutation accumulation. In addition to alkylating agents, the MMR pathway may also induce a response to other agents such as 6-thioguanine (73,74), 5-fluorouracil (75–77), and the intrastrand crosslinker cisplatin (78–81), though the mechanism of response to these drugs is less well understood.
4.2 Response of MMR-defective cancers to chemotherapy
The discovery of the MMR-dependent damage response is clinically relevant as it pertains to tumor response to chemotherapy. Studies examining the benefits of 5-fluorouracil (5-FU) in patients with MSI+ colorectal cancer concluded that the therapy had no effect on overall survival or disease free survival, while benefits were observed in patients with microsatellite stable tumors (82–87) (also see Li, Pearlman & Hsieh in this issue). This clinical result is consistent with cellular studies observing enhanced drug resistance in MMR-defective cells. For 5-FU specifically, laboratory studies of human cancer cell lines demonstrated that 5-FU was more effective in MMR-proficient cells as treatment leads to an incorporation of 5-fluoro-2’-deoxyuridine into DNA. These lesions are commonly mispaired with guanine (75–77) and serve as a substrate for the MMR proteins likely resulting in a similar damage response mechanism as observed with MeG-T mismatches (77). 5-FU also increases levels of deoxyuridine by decreasing thymidine nucleotide pools. Deoxyuridine misincorporation into DNA results in formation of U-G mispairs that also stimulate MMR activity (77). Thus, treatment of cancer cells with 5-FU may cause multiple lesions that activate a MMR-dependent damage response.
Preclinical studies have also demonstrated increased resistance to cisplatin and carboplatin in MMR-defective cells (78–81) (also see Li, Pearlman & Hsieh and Begum & Martin in this issue). The MMR pathway recognizes primarily GpG intrastrand crosslinks that are generated by platinum-containing agents. These studies may explain clinical observations of MMR loss in recurrent ovarian tumors following treatment with cisplatin (79,88–92). Treatment of the primary tumor with cisplatin may lead to selection for a subset of cancer cells that have lost MMR function. A similar observation was made in gliomas following treatment with the alkylating agent temozolomide as increased mutations in MSH6 were found in recurrent tumors that were not present in the primary cancer (93,94).
As noted earlier, MMR-defective sporadic cancers mostly arise due to hypermethylation of the MLH1 promoter, thus basic science studies have examined the effects of demethylating agents such as 2’deoxy-5-azacytidine (decitabine) on the ability to restore MLH1 expression, and therefore function. Decitabine treatment of an ovarian cancer cell line was shown to restore MLH1 expression as well as increase sensitivity to cisplatin, carboplatin and temozolomide (95,96). This effect was enhanced in cells treated with decitabine and the histone deacetylase inhibitor belinostat (97). These studies spurred clinical trials examining combinations of decitabine and other chemotherapies. A phase II clinical trial examining decitabine pretreatment followed by carboplatin suggests that the combination improves response rate and progression free survival (98). Similarly, chemotherapeutic resistance of metastatic melanoma to temozolomide appears improved by treatment with decitabine (99).
5. A peek into the lab to see what is next
5.1 Synthetic lethality and cancer treatment
An emerging area of cancer research that holds promise for new therapeutic approaches involves the exploitation of synthetic lethal interactions between DNA damage response pathways, including MMR (100,101) (also see Begum & Martin in this issue). Synthetic lethal interactions occur between two genes or pathways when the loss of both leads to cell death, whereas the loss of either one alone is tolerated. The best characterized synthetic lethal relationship involving a DNA damage response pathway involves the chemical inhibition of poly(ADP-ribose) polymerase 1 (PARP1) in cells deficient for the homologous recombination repair (HRR) proteins BRCA1 or BRCA2 (102,103). PARP1 responds to single-stranded DNA breaks and prevents their degradation into toxic double strand breaks by either stabilizing the lesion or recruiting other DNA proteins to repair the break (104). Increased conversion of these lesions into double strand breaks in the absence of PARP1 activity would be particularly toxic to cells defective in homologous recombination repair. Recently, the PARP1 inhibitor olaparib has been approved by the US Food and Drug Administration for use in cancers with defects in the BRCA genes and is being explored for use in cancers with other defects in the HRR pathway (100).
Synthetic lethal interactions involving the MMR pathway were first described in yeast with mutants in pol3 that affect its proofreading exonuclease activity (105–107). Cell death may occur due to a possible mutational overload resulting from loss of both error correction functions. Synthetic lethal interactions with MMR in human cells were identified through small interfering RNA (siRNA) screens in MSH2 or MLH1 defective cell lines (108). As described in more detail in the review by Begum & Martin in this issue, the knockdown of DNA polymerase β in MSH2 deficient cells results in increased cell death. Likewise, MLH1 defective cells were sensitive to knockdown of DNA polymerase γ, both polymerases thought to be involved in the repair of 8-oxo-guanine lesions. One rationale for these findings comes from earlier studies that showed that MMR proteins can bind to 8-oxo-guanine-adenine mispairs in vitro (109,110) and that MSH2 deficient cells are more sensitive to treatment with the oxidizing agent methotrexate (111) (also see review by Crouse in this issue). Thus, the MMR pathway may play an important back-up role in the repair of oxidative damage in DNA, a function that can be exploited in MMR-defective cells. A similar siRNA screen against an array of cellular kinases and associated proteins determined that MSH2, MLH1 and MSH6 were also synthetically lethal with the PTEN-induced putative kinase 1 (PINK1) gene (112). A corresponding accumulation of 8-oxo-guanine lesions in these cells suggested that loss of PINK1, a mitochondrial kinase that regulates cytochrome c release from the mitochondria, may affect mitochondrial membrane potential leading to increased oxidative stress. Therefore, therapeutic strategies that increase the levels of 8-oxo-guanine lesions in MMR-defective cancers cells may exploit the lack of a MMRdependent clearing of these lesions, resulting in tumor specific cell death. From these studies, one may hypothesize that another potential target for synthetic lethality with MMR is MTH1 which plays a key role in removing oxidized dNTPs from cells to prevent their incorporation into DNA. Recent studies have shown that cancer cells are addicted to the activity of MTH1 and that inhibitors of this enzyme can selectively kill cancer cells (113). MMR defective cancers may be particularly susceptible to MTH1 inhibitors.
5.2 Mutated genes in MMR-defective cancers
Understanding the mechanism by which defects in MMR cause cancer may guide new treatment or prevention strategies. Loss of MMR has been shown to increase mutation frequency several hundred fold and this mutator phenotype has been proposed to increase the chances of acquiring mutation in other important growth control or survival genes (114). In particular, loss of MMR has been associated with an increase of frameshift mutations in small sequence repeats within the coding regions of genes (115–118). The first observation of this mutator phenotype at work involved the detection of a frameshift mutation in a run of adenines within the coding region of the TGFβ type II receptor gene in MMR-defective cancer cells (116). The frameshift leads to a premature stop codon and production of a truncated protein product which abrogates the TGFβ signaling pathway. Since that first discovery, a number of other genes have been identified that display increased mutation within intra-genic repeats. Many of these genes had not been previously implicated in colorectal tumorigenesis suggesting that the pathway to tumor development may differ in MMR-defective cancers. Consistent with this idea, exome sequencing of colorectal carcinomas has revealed a subset of cancers that display a hypermutator phenotype marked by a set of commonly mutated genes that differ from those altered in non-hypermutated tumors (119). Of the hypermutated tumors, 77% were marked by MSI with the majority of those containing a hypermethylated MLH1. Thus, basic science studies, particularly increased genome sequencing studies of MMR-defective cancers, may identify new targets for therapeutic intervention in these tumors. One cautionary note, however, is that due to the hypermutability of these tumors, many of the mutations identified may merely be passenger mutations that do not directly contribute to tumor phenotype. Also, it is not entirely clear how similar the development of sporadic MMR-deficient tumors is to LS cancers. Sporadic MMR-deficient tumors are more commonly marked by enhanced CpG island methylation which may lead to downregulation of subsets of genes not affected in LS (120–122). Sporadic MSI+ tumors are also associated with oncogenic BRAF mutations, which are rarely found in LS tumors (122,123). Due to this fact, testing for BRAF mutations is sometimes used clinically to distinguish whether a cancer is likely sporadic versus LS-associated (124–126) (also see Sijmons and Hofstra in this issue).
6. Conclusions
The buzzwords personalized medicine, precision medicine and genomic medicine are frequently used these days by researchers and clinicians to describe a new world where medical treatment will be more tailored to individuals based on specific genetic or molecular biomarkers. However, this approach has already been utilized in the LS field for over a decade. The combination of a well-defined disease caused by mutations in a well-researched cellular pathway has led to a wealth of information that has aided patients and family members suffering from this condition. The story of LS provides a hopeful model for the use of molecular and genetic information to precisely diagnose and treat other diseases. At the same time, the story of LS underscores the absolute necessity of continued investment in basic science research – from bacteria to humans – if this vision of improved personalized medical care is to become a reality.
Highlights.
The history of LS and its association with MMR is discussed
Basic science research has greatly informed the clinical management of LS
Diagnosis has been aided by understanding the molecular biology of faulty MMR
Laboratory studies have also offered clues about tumor response to therapy
Strong basic science research will drive the success of precision medicine efforts
Acknowledgements
This work was supported by the National Institutes of Health CA181959 and the State of Connecticut Grant 13SCB-UCHC-06.
Abbreviations
- LS
Lynch syndrome
- CRC
colorectal cancer
- HNPCC
hereditary non-polyposis colon cancer
- MMR
mismatch repair
- FAP
familial adenomatous polyposis
- CFS
cancer family syndrome
- MSI
microsatellite instability
- IDL
insertion/deletion loop
- MSH2
human mutS homolog 2
- MLH1
human mutL homolog 1
- MSH6
human mutS homolog 6
- PMS2
post meiotic segregation increased 2
- IHC
immunohistochemistry
- VUS
variants of uncertain significance
- MNNG
N-methyl-N’-nitro-N-nitrosoguanidine
- MeG
O6-methylguanine
- MGMT
O6-methylguanine-DNA methyltransferase
- 5-FU
5-fluorouracil
- PARP1
poly(ADP-ribose) polymerase 1
- BRCA1
breast cancer 1 early onset
- BRCA2
breast cancer 2 early onset
- HRR
homologous recombination repair
- siRNA
small interfering RNA
- PINK1
PTEN-induced putative kinase 1
- MTH1
human mutT homolog 1
- TGFβ1
transforming growth factor beta 1
- BRAF
v-raf murine sarcoma viral oncogene homolog B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lynch H, Lynch P, Lanspa S, Snyder C, Lynch J, Boland C. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clinical Genetics. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP. Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer. 2015;15:181–194. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 3.Boland CR, Lynch H. The History of Lynch Syndrome. Familial Cancer. 2013;12:145–157. doi: 10.1007/s10689-013-9637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warthin A. Hereditary with reference to carcinoma as shown by the study of the cases examined in the Pathological Laboratory of the University of Michigan, 1895–1912. Arch Int Med. 1913;12:546–555. [Google Scholar]
- 5.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 6.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 7.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer [see comments] Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 8.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 9.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 10.Farabaugh PJ, Schmeissner U, Hofer M, Miller JH. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978;126:847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- 11.Streisinger G, Owen J. Mechanisms of spontaneous and induced frameshift mutation in bacteriophage T4. Genetics. 1985;109:633–659. doi: 10.1093/genetics/109.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrich P. Methyl-directed DNA mismatch correction. J. Biol. Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- 13.Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 14.Parsons R, Li G-M, Longley MJ, Fang W-h, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 15.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 16.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 17.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 18.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 19.Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 20.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 21.Worthley DL, Walsh MD, Barker M, Ruszkiewicz A, Bennett G, Phillips K, Suthers G. Familial Mutations in PMS2 Can Cause Autosomal Dominant Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 2005;128:1431–1436. doi: 10.1053/j.gastro.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 22.Lynch HT, Lynch PM. Molecular Screening for the Lynch Syndrome -- Better Than Family History? N Engl J Med. 2005;352:1920–1922. doi: 10.1056/NEJMe058058. [DOI] [PubMed] [Google Scholar]
- 23.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 24.Umar A, Boland CR, Terdiman JP, Syngal S, Chapelle Adl, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HFA, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN. Immunohistochemistry Versus Microsatellite Instability Testing in Phenotyping Colorectal Tumors. J Clin Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 26.Byfield SAD, Syngal S. Clinical Guidelines Versus Universal Molecular Testing: Are We Ready to Choose an Optimal Strategy for Lynch Syndrome Identification[quest] Am J Gastroenterol. 2008;103:2837–2840. doi: 10.1111/j.1572-0241.2008.02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A. Screening for the Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 28.Julie C, Tresallet C, Brouquet A, Vallot C, Zimmermann U, Mitry E, Radvanyi F, Rouleau E, Lidereau R, Coulet F, Olschwang S, Frebourg T, Rougier P, Nordlinger B, Laurent-Puig P, Penna C, Boileau C, Franc B, Muti C, Hofmann-Radvanyi H. Identification in Daily Practice of Patients With Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer): Revised Bethesda Guidelines- Based Approach Versus Molecular Screening. Am J Gastroenterol. 2008;103:2825–2835. doi: 10.1111/j.1572-0241.2008.02084.x. [DOI] [PubMed] [Google Scholar]
- 29.Morrison J, Bronner M, Leach BH, Downs-Kelly E, Goldblum JR, Liu X. Lynch syndrome screening in newly diagnosed colorectal cancer in general pathology practice: From the revised Bethesda guidelines to a universal approach. Scandinavian Journal of Gastroenterology. 2011;46:1340–1348. doi: 10.3109/00365521.2011.610003. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, Alenda C, Payá A, Brea A, Egoavil CM, Castillejo A, Barberá VM, Bessa X, Xicola RM, Rodríguez-Soler M, Sánchez-Fortún C, Acame N, Castellví-Bel S, Piñol V, Balaguer F, Bujanda L, De-Castro M-L, Llor X, Andreu M, Carracedo A, Soto J-L, Castells A, Jover R. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61:865–872. doi: 10.1136/gutjnl-2011-300041. [DOI] [PubMed] [Google Scholar]
- 31.Cunningham J, Christensen E, Tester D, Kim C, Roche P, Burgart L, Thibodeau S. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 32.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa J-PJ, Markowitz S, Willson JKV, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. PNAS. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kane M, Loda M, Gaida G, Lipman J, Mishra R, Goldman H, Jessup J, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 34.Plazzer JP, Sijmons RH, Woods MO, Peltomaki P, Thompson B, Den Dunnen JT, Macrae F. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer. 2013;12:175–180. doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 35.Heinen CD, Rasmussen LJ. Determining the functional significance of mismatch repair gene missense variants using biochemical and cellular assays. Hereditary Cancer in Clinical Practice. 2012;10:9. doi: 10.1186/1897-4287-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasmussen LJ, Heinen CD, Royer-Pokora B, Drost M, Tavtigian S, Hofstra RMW, de Wind N. Pathological assessment of mismatch repair gene variants in Lynch syndrome: Past, present, and future. Human Mutation. 2012;33:1617–1625. doi: 10.1002/humu.22168. [DOI] [PubMed] [Google Scholar]
- 37.Thompson BA, Spurdle AB, Plazzer J-P, Greenblatt MS, Akagi K, Al-Mulla F, Bapat B, Bernstein I, Capella G, den Dunnen JT, du Sart D, Fabre A, Farrell MP, Farrington SM, Frayling IM, Frebourg T, Goldgar DE, Heinen CD, Holinski-Feder E, Kohonen-Corish M, Robinson KL, Leung SY, Martins A, Moller P, Morak M, Nystrom M, Peltomaki P, Pineda M, Qi M, Ramesar R, Rasmussen LJ, Royer-Pokora B, Scott RJ, Sijmons R, Tavtigian SV, Tops CM, Weber T, Wijnen J, Woods MO, Macrae F, Genuardi M on behalf of InSiGHT. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes J, Jr, Clark S, Modrich P. Strand-Specific Mismatch Correction in Nuclear Extracts of Human and Drosophila melanogaster Cell Lines. PNAS. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human Mismatch Repair: Reconstitution of a Nick-Directed Bidirectional Reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson A, Gu L, Li GM. Reconstitution of 5'-Directed Human Mismatch Repair in a Purified System. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Drost M, Zonneveld JBM, van Hees S, Rasmussen LJ, Hofstra RMW, de Wind N. A rapid and cell-free assay to test the activity of Lynch syndrome-associated MSH2 and MSH6 missense variants. Human Mutation. 2012;33:488–494. doi: 10.1002/humu.22000. [DOI] [PubMed] [Google Scholar]
- 42.Drost M, Zonneveld JéBM, van Dijk L, Morreau H, Tops CM, Vasen HFA, Wijnen JT, de Wind N. A cell-free assay for the functional analysis of variants of the mismatch repair protein MLH1. Human Mutation. 2010;31:247–253. doi: 10.1002/humu.21180. [DOI] [PubMed] [Google Scholar]
- 43.Kariola R, Raevaara TE, Lonnqvist KE, Nystrom-Lahti M. Functional analysis of MSH6 mutations linked to kindreds with putative hereditary non-polyposis colorectal cancer syndrome. Hum. Mol. Genet. 2002;11:1303–1310. doi: 10.1093/hmg/11.11.1303. [DOI] [PubMed] [Google Scholar]
- 44.Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D'Urso M, Venuta S, Jiricny J. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33:160–167. [PubMed] [Google Scholar]
- 45.Ollila S, Dermadi Bebek D, Jiricny J, Nystrom M. Mechanisms of pathogenicity in human MSH2 missense mutants. Hum Mutat. 2008;29:1355–1363. doi: 10.1002/humu.20893. [DOI] [PubMed] [Google Scholar]
- 46.Guerrette S, Wilson T, Gradia S, Fishel R. Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer. Mol Cell Biol. 1998;18:6616–6623. doi: 10.1128/mcb.18.11.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinen CD, Wilson T, Mazurek A, Berardini M, Butz C, Fishel R. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–478. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 48.Lutzen A, de Wind N, Georgijevic D, Nielsen F, Rasmussen L. Functional analysis of HNPCC-related missense mutations in MSH2. Mutat Res. 2008;645:44–55. doi: 10.1016/j.mrfmmm.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 49.Ollila S, Sarantaus L, Kariola R, Chan P, Hampel H, Holinski-Feder E, Macrae F, Kohonen-Corish M, Gerdes A-M, Peltomäki P, Mangold E, de la Chapelle A, Greenblatt M, Nyström M. Pathogenicity of MSH2 Missense Mutations Is Typically Associated With Impaired Repair Capability of the Mutated Protein. Gastroenterology. 2006;131:1408–1417. doi: 10.1053/j.gastro.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 50.Perera S, Bapat B. The MLH1 variants p.Arg265Cys and p.Lys618Ala affect protein stability while p.Leu749Gln affects heterodimer formation. Human Mutation. 2008;29:332–332. doi: 10.1002/humu.9523. [DOI] [PubMed] [Google Scholar]
- 51.Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 52.Jager AC, Rasmussen M, Bisgaard HC, Singh KK, Nielsen FC, Rasmussen LJ. HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene. 2001;20:3590–3595. doi: 10.1038/sj.onc.1204467. [DOI] [PubMed] [Google Scholar]
- 53.Raevaara TE, Korhonen M, H L, H H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, W M, S D, Gerdes A, Peltomaki P, Kohonen-Corish M, Mangold E, Macrae F, M G, de la Chapelle A, M N. Functional Significance and Clinical Phenotype of Nontruncating Mismatch Repair Variants of MLH1. Gastroenterology. 2005;129:537–549. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Mastrocola AS, Heinen CD. Lynch syndrome-associated mutations in MSH2 alter DNA repair and checkpoint response functions in vivo. Human Mutation. 2010;31:E1699–E1708. doi: 10.1002/humu.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wielders EAL, Dekker RJ, Holt I, Morris GE, te Riele H. Characterization of MSH2 variants by endogenous gene modification in mouse embryonic stem cells. Human Mutation. 2011;32:389–396. doi: 10.1002/humu.21448. [DOI] [PubMed] [Google Scholar]
- 56.Wielders EAL, Houlleberghs H, Isik G, te Riele H. Functional Analysis in Mouse Embryonic Stem Cells Reveals Wild-Type Activity for Three Msh6 Variants Found in Suspected Lynch Syndrome Patients. PLoS ONE. 2013;8:e74766. doi: 10.1371/journal.pone.0074766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drost M, Lützen A, van Hees S, Ferreira D, Calléja F, Zonneveld JBM, Nielsen FC, Rasmussen LJ, de Wind N. Genetic screens to identify pathogenic gene variants in the common cancer predisposition Lynch syndrome. Proceedings of the National Academy of Sciences. 2013;110:9403–9408. doi: 10.1073/pnas.1220537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinen CD, Schmutte C, Fishel R. DNA Repair and Tumorigenesis: Lessons from Hereditary Cancer Syndromes. Cancer Biology & Therapy. 2002;1:477–485. doi: 10.4161/cbt.1.5.160. [DOI] [PubMed] [Google Scholar]
- 59.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mechanisms of Ageing and Development. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 61.Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, doublestrand breaks, cell proliferation and signaling. Biochemical Pharmacology. 2003;66:1547–1554. doi: 10.1016/s0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 62.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 63.Kat A, Thilly WG, Fang WH, Longley MJ, Li GM, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci U S A. 1993;90:6424–6428. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fahrer J, Kaina B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis. 2013;34:2435–2442. doi: 10.1093/carcin/bgt275. [DOI] [PubMed] [Google Scholar]
- 65.Cejka P, Stojic L, Mojas N, Russell AM, Heinimann K, Cannavo E, di Pietro M, Marra G, Jiricny J. Methylation-induced G2/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaina B, Ziouta A, Ochs K, Coquerelle T. Chromosomal instability, reproductive cell death and apoptosis induced by O6-methylguanine in Mex-, Mex+ and methylation-tolerant mismatch repair compromised cells: facts and models. Mutat Res. 1997;381:227–241. doi: 10.1016/s0027-5107(97)00187-5. [DOI] [PubMed] [Google Scholar]
- 67.Mojas N, Lopes M, Jiricny J. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adamson A, Beardsley D, Kim W, Gao Y, Baskaran R, Brown K. Methylator-induced, Mismatch Repair-dependent G2 Arrest Is Activated through Chk1 and Chk2. Molecular Biology of the Cell. 2005;16:1513–1526. doi: 10.1091/mbc.E04-02-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Interactions of Human Mismatch Repair Proteins MutS and MutL with Proteins of the ATR-Chk1 Pathway. Journal of Biological Chemistry. 2010;285:5974–5982. doi: 10.1074/jbc.M109.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noonan EM, Shah D, Yaffe MB, Lauffenburger DA, Samson LD. O 6-Methylguanine DNA lesions induce an intra-S-phase arrest from which cells exit into apoptosis governed by early and late multi-pathway signaling network activation. Integrative Biology. 2012;4:1237–1255. doi: 10.1039/c2ib20091k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Qin J. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. PNAS. 2003;100:15387–15392. doi: 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshioka K, Yoshioka Y, Hsieh P. ATR Kinase Activation Mediated by MutSa and MutLa in Response to Cytotoxic O6-Methylguanine Adducts. Mol Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hawn MT, Umar A, Carethers JM, Marra G, Kunkel TA, Boland CR, Koi M. Evidence for a Connection between the Mismatch Repair System and the G2 Cell Cycle Checkpoint. Cancer Research. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 74.Swann PF, Waters TR, Moulton DC, Xu YZ, Zheng Q, Edwards M, Mace R. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 75.Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. International Journal of Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- 76.Carethers JM, Chauhan DP, Daniel F, Sibylle N, Robert SB, Howell Stephen B, Boland C.Richard. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA Mismatch Repair-dependent Response to Fluoropyrimidine-generated Damage. J. Biol. Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 78.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen RD, Boland CR, Koi M, Fishel R, Howell SB. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 79.Brown R, Hirst GL, Gallagher WM, McIlwrath AJ, Margison GP, van der Zee AG, Anthoney DA. hMLH1 expression and cellular responses of ovarian tumour cells to treatment with cytotoxic anticancer agents. Oncogene. 1997;15:45–52. doi: 10.1038/sj.onc.1201167. [DOI] [PubMed] [Google Scholar]
- 80.Drummond JT, Anthoney A, Brown R, Modrich P. Cisplatin and Adriamycin Resistance Are Associated with MutLα and Mismatch Repair Deficiency in an Ovarian Tumor Cell Line. Journal of Biological Chemistry. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 81.Fink D, Nebel S, Aebi S, Zheng H, Cenni B, Nehme A, Christen RD, Howell SB. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 82.Benatti P, Gafa R, Barana D, Marino M, Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L, Menigatti M, Roncari B, Maffei S, Rossi G, Ponti G, Santini A, Losi L, Di Gregorio C, Oliani C, Ponz de Leon M, Lanza G. Microsatellite Instability and Colorectal Cancer Prognosis. Clin Cancer Res. 2005;11:8332–8340. doi: 10.1158/1078-0432.CCR-05-1030. [DOI] [PubMed] [Google Scholar]
- 83.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5- fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 84.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Pinol V, Xicola RM, Bujanda L, Rene JM, Clofent J, Bessa X, Morillas JD, Nicolas-Perez D, Paya A, Alenda C. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy on colorectal cancer. Gut. 2006;55:848–855. doi: 10.1136/gut.2005.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor Microsatellite-Instability Status as a Predictor of Benefit from Fluorouracil-Based Adjuvant Chemotherapy for Colon Cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz J-F, Sinicrope F, Gallinger S. Defective Mismatch Repair As a Predictive Marker for Lack of Efficacy of Fluorouracil-Based Adjuvant Therapy in Colon Cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, Yothers G, Allegra C, Moore MJ, Gallinger S, Sargent DJ. DNA Mismatch Repair Status and Colon Cancer Recurrence and Survival in Clinical Trials of 5-Fluorouracil-Based Adjuvant Therapy. Journal of the National Cancer Institute. 2011;103:863–875. doi: 10.1093/jnci/djr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fink D, Nebel S, Norris PS, Baergen RN, Wilczynski SP, Costa MJ, Haas M, Cannistra SA, Howell SB. Enrichment for DNA mismatch repair-deficient cells during treatment with cisplatin. International Journal of Cancer. 1998;77:741–746. doi: 10.1002/(sici)1097-0215(19980831)77:5<741::aid-ijc13>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 89.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The Acquisition of hMLH1 Methylation in Plasma DNA after Chemotherapy Predicts Poor Survival for Ovarian Cancer Patients. Clinical Cancer Research. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 90.Mayer F, Gillis AJM, Dinjens W, Oosterhuis JW, Bokemeyer C, Looijenga LHJ. Microsatellite Instability of Germ Cell Tumors Is Associated with Resistance to Systemic Treatment. Cancer Research. 2002;62:2758–2760. [PubMed] [Google Scholar]
- 91.Samimi G, Fink D, Varki NM, Husain A, Hoskins WJ, Alberts DS, Howell SB. Analysis of MLH1 and MSH2 Expression in Ovarian Cancer before and after Platinum Drug-based Chemotherapy. Clinical Cancer Research. 2000;6:1415–1421. [PubMed] [Google Scholar]
- 92.Watanabe Y, Koi M, Hemmi H, Hoshai H, Noda K. A change in microsatellite instability caused by cisplatin-based chemotherapy of ovarian cancer. Br J Cancer. 2001;85:1064–1069. doi: 10.1054/bjoc.2001.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, Iafrate AJ, Louis DN. Loss of the Mismatch Repair Protein MSH6 in Human Glioblastomas Is Associated with Tumor Progression during Temozolomide Treatment. Clinical Cancer Research. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of Drug Resistance in Human Tumor Xenografts by 2'-Deoxy-5-azacytidine-induced Demethylation of the hMLH1 Gene Promoter. Cancer Res. 2000;60:6039–6044. [PubMed] [Google Scholar]
- 96.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 97.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer. 2009;100:758–763. doi: 10.1038/sj.bjc.6604932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic Resensitization to Platinum in Ovarian Cancer. Cancer Research. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tawbi HA, Beumer JH, Tarhini AA, Moschos S, Buch SC, Egorin MJ, Lin Y, Christner S, Kirkwood JM. Safety and efficacy of decitabine in combination with temozolomide in metastatic melanoma: a phase I/II study and pharmacokinetic analysis. Annals of Oncology. 2013;24:1112–1119. doi: 10.1093/annonc/mds591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 101.Martin SA, Lord CJ, Ashworth A. Therapeutic Targeting of the DNA Mismatch Repair Pathway. Clin. Cancer Res. 2010;16:5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- 102.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 103.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NMB, Jackson SP, Smith GCM, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 104.Woodhouse BC, Dianov GL. Poly ADP-ribose polymerase-1: An international molecule of mystery. DNA Repair. 2008;7:1077–1086. doi: 10.1016/j.dnarep.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 105.Argueso JL, Smith D, Yi J, Waase M, Sarin S, Alani E. Analysis of Conditional Mutations in the Saccharomyces cerevisiae MLH1 Gene in Mismatch Repair and in Meiotic Crossing Over. Genetics. 2002;160:909–921. doi: 10.1093/genetics/160.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Molecular and Cellular Biology. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. DNA Polymerases as Potential Therapeutic Targets for Cancers Deficient in the DNA Mismatch Repair Proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–248. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mazurek A, Berardini M, Fishel R. Activation of Human MutS Homologs by 8-Oxo-guanine DNA Damage. J. Biol. Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- 110.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 111.Martin SA, McCarthy A, Barber LJ, Burgess DJ, Parry S, Lord CJ, Ashworth A. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Molecular Medicine. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. Parallel High-Throughput RNA Interference Screens Identify PINK1 as a Potential Therapeutic Target for the Treatment of DNA Mismatch Repair–Deficient Cancers. Cancer Research. 2011;71:1836–1848. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]
- 113.Gad H, Koolmeister T, Jemth A-S, Eshtad S, Jacques SA, Strom CE, Svensson LM, Schultz N, Lundback T, Einarsdottir BO, Saleh A, Gokturk C, Baranczewski P, Svensson R, Berntsson RPA, Gustafsson R, Stromberg K, Sanjiv K, Jacques-Cordonnier M-C, Desroses M, Gustavsson A-L, Olofsson R, Johansson F, Homan EJ, Loseva O, Brautigam L, Johansson L, Hoglund A, Hagenkort A, Pham T, Altun M, Gaugaz FZ, Vikingsson S, Evers B, Henriksson M, Vallin KSA, Wallner OA, Hammarstrom LGJ, Wiita E, Almlof I, Kalderen C, Axelsson H, Djureinovic T, Puigvert JC, Haggblad M, Jeppsson F, Martens U, Lundin C, Lundgren B, Granelli I, Jensen AJ, Artursson P, Nilsson JA, Stenmark P, Scobie M, Berglund UW, Helleday T. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature. 2014;508:215–221. doi: 10.1038/nature13181. [DOI] [PubMed] [Google Scholar]
- 114.Fishel R, Kolodner RD. Identification of mismatch repair genes and their role in the development of cancer. Curr Opin Genet Dev. 1995;5:382–395. doi: 10.1016/0959-437x(95)80055-7. [DOI] [PubMed] [Google Scholar]
- 115.Duval A, Hamelin R. Mutations at Coding Repeat Sequences in Mismatch Repair-deficient Human Cancers: Toward a New Concept of Target Genes for Instability. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 116.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, Brattain M, Willson JKV. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 117.Mori Y, Yin J, Rashid A, Leggett BA, Young J, Simms L, Kuehl PM, Langenberg P, Meltzer SJ, Stine OC. Instabilotyping: Comprehensive Identification of Frameshift Mutations Caused by Coding Region Microsatellite Instability. Cancer Research. 2001;61:6046–6049. [PubMed] [Google Scholar]
- 118.Percesepe A, Kristo P, Aaltonen LA, Ponz de Leon M, de la Chapelle A, Peltomaki P. Mismatch repair genes and mononucleotide tracts as mutation targets in colorectal tumors with different degrees of microsatellite instability. Oncogene. 1998;17:157–163. doi: 10.1038/sj.onc.1201944. [DOI] [PubMed] [Google Scholar]
- 119.CGA Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Toyota M, Issa J-PJ. CpG island methylator phenotypes in aging and cancer. Seminars in Cancer Biology. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- 121.Issa J-P. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 122.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 123.Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151–157. doi: 10.1136/jmedgenet-2011-100714. [DOI] [PubMed] [Google Scholar]
- 124.Bouzourene H, Hutter P, Losi L, Martin P, Benhattar J. Selection of patients with germline MLH1 mutated Lynch syndrome by determination of MLH1 methylation and BRAF mutation. Familial Cancer. 2010;9:167–172. doi: 10.1007/s10689-009-9302-4. [DOI] [PubMed] [Google Scholar]
- 125.Jensen LH, Lindebjerg J, Byriel L, Kolvraa S, Crüger DG. Strategy in clinical practice for classification of unselected colorectal tumours based on mismatch repair deficiency. Colorectal Disease. 2008;10:490–497. doi: 10.1111/j.1463-1318.2007.01378.x. [DOI] [PubMed] [Google Scholar]
- 126.Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Familial Cancer. 2007;6:301–310. doi: 10.1007/s10689-007-9124-1. [DOI] [PubMed] [Google Scholar]