Fig 6. Loss of endonuclease motifs in E. coli MutL.
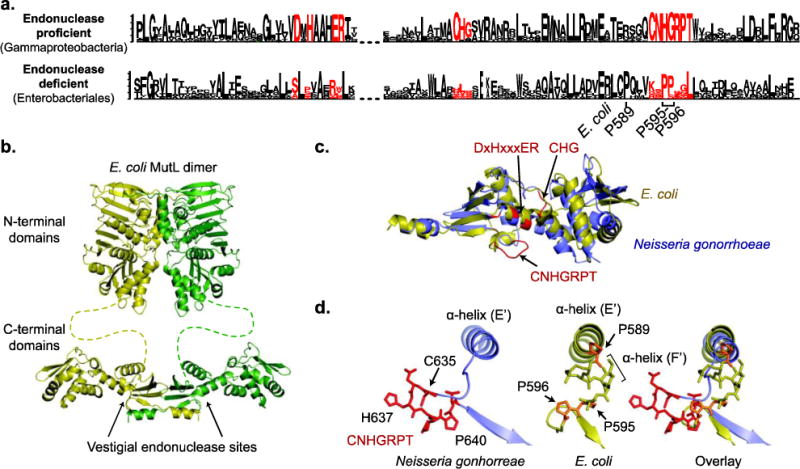
a. Sequence logos, where the height of the letter indicates of its degree of conservation, were generated by Seq2Logo [107] for the endonuclease-proficient MutL in Gammaproteobacteria and the endonuclease-deficient MutL in Enterobacteriales, which includes E. coli. The endonuclease motifs are shown in red. b. Modeled structure of full length MutL based on the N-terminal domain structure (PDB id 1b62; [108]) and the C-terminal domain structure (PDB id 1×9z; [57]). c. Overlay of the E. coli (yellow) and Neisseria gonorrhoeae (green) MutL C-terminal domains reveals that the folds are the same; N. gonorrhoeae is the nuclease-proficient domain structure that is most closely related to E. coli (Fig. 1). Residues in red correspond to the endonuclease motifs. d. Changes in the CNHGRPT motif-containing loop are depicted for the nuclease-proficient N. gonorrhoeae (left) and E. coli (middle) with an overlay of the two structures (right).
