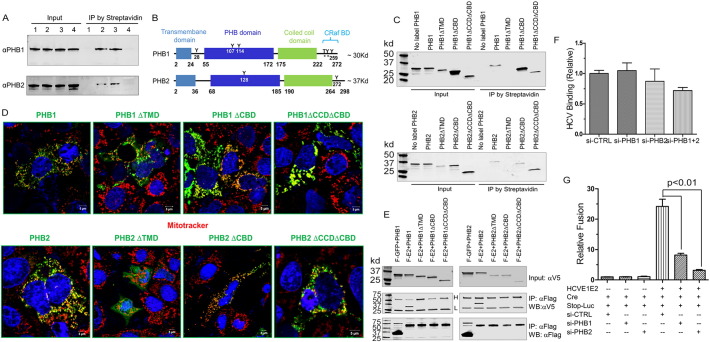
Fig. 2.
PHB1 and 2 mediate HCV entry via a novel mechanism.
(A) Huh7.5.1 cells were surface biotinylated as described in the Materials and Methods section. IP was done using streptavidin agarose beads. The presence of PHB1 and PHB2 was detected by Western blotting. Lane 1, no biotin labeling; lane 2, biotin labeled; lane 3, HCVcc infected cells labeled with biotin; lane 4, RocA (20 nM) treated cells labeled with biotin. Of note, because Roc-A treatment decreased the total cellular PHB1 and 2 (Shown in Fig. 3B), we intentionally used twice as many cells as starting materials for the Roc-A treated group (Lane 4) in order to achieve similar amount of prohibitins in the input in comparison to the levels in other lanes.
(B) Domain organization of PHBs.
(C) Surface biotinylation assay on individual PHB deletion mutants.
(D) Confocal images of PHB deletion mutants. Green, PHB mutants; Red, mitotracker. (E), co-IP studies of Flag-E2 (F-E2) and V5-tagged PHB deletion mutants.
(F) Real-time quantification of HCV RNA bound to cells in which PHB1 or PHB2 were knocked down. Results were calculated as relative RNA copies with numbers obtained from si-CTRL transfected cells set to 1 (mean ± SD, *p < 0.05).
(G) 293 T-CLDN1 cells expressing HCV E1E2 and Cre were fused to Huh7.5.1 cells that were transfected with siRNA and Stop-Luc expressing plasmid. Luciferase activity was determined 24 h after mixing (mean ± SD, *p < 0.05).