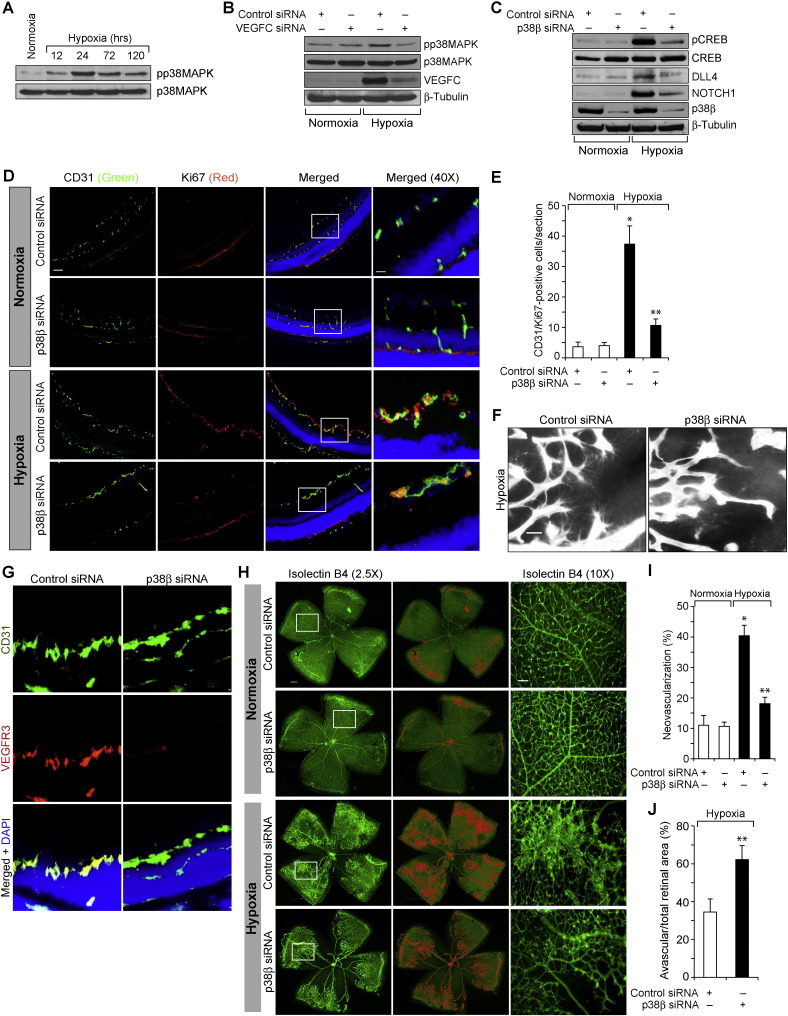
Fig. 9.
p38MAPKβ mediates hypoxia-induced CREB–DLL4–NOTCH1 activation and neovascularization. A. An equal amount of protein from normoxic and various time periods of hypoxic retinal extracts were analyzed by Western blotting for pp38MAPK levels using its phospho-specific antibodies and normalized to its total level. B & C. All the conditions were the same as in panel A except that pups were administered intravitreally with 1 μg/0.5 μl/eye of scrambled, VEGFC, or p38β siRNA at P10 and P11 and at P13 the retinas were isolated and tissue extracts were prepared. An equal amount of protein from normoxic and hypoxic retinal extracts were analyzed by Western blotting for the indicated proteins using their specific antibodies and normalized to β-tubulin. D. All the conditions were the same as in panel C except that pups were administered intravitreally with 1 μg/0.5 μl/eye of scrambled, or p38β siRNA at P12 and P13 and at P15 retinas were isolated, cross-sections were made and stained by immunofluorescence for CD31 and Ki67. E. Retinal EC proliferation was measured by counting CD31- and Ki67-positive cells that extended anterior to the inner limiting membrane per section (n = 6 eyes, 3 sections/eye). F. All the conditions were the same as in panel C except that that pups were administered intravitreally with 1 μg/0.5 μl/eye of scrambled, or p38β siRNA at P12, P13 and P15 and at P17 the retinas were isolated, stained with isolectin B4 and flat mounts were made and examined for filopodia formation. G. All the conditions were the same as in panel D except that the sections were stained for CD31, VEGFR3 and DAPI. H. All the conditions were the same as in panel F except that the flat mounts were examined for neovascularization. Retinal vascularization is shown in the first column. Neovascularization is highlighted in red in the second column. The third column shows the selected rectangular areas of the images shown in the first column under 10X magnification. I & J. Retinal neovascularization (I) and avascular areas (J) were determined as described in “Materials and Methods.” The bar graphs represent quantitative analysis of 6 retinas. The values are presented as Mean ± SD. * p < 0.01 vs normoxia, normoxia + control siRNA; ** p < 0.01 vs hypoxia + control siRNA. Scale bar represents 50 μm and 20 μm in panel D, far left column and far right column, respectively, 20 μm in panels F & G and 300 μm and 50 μm in panel H, far left column and far right column, respectively.