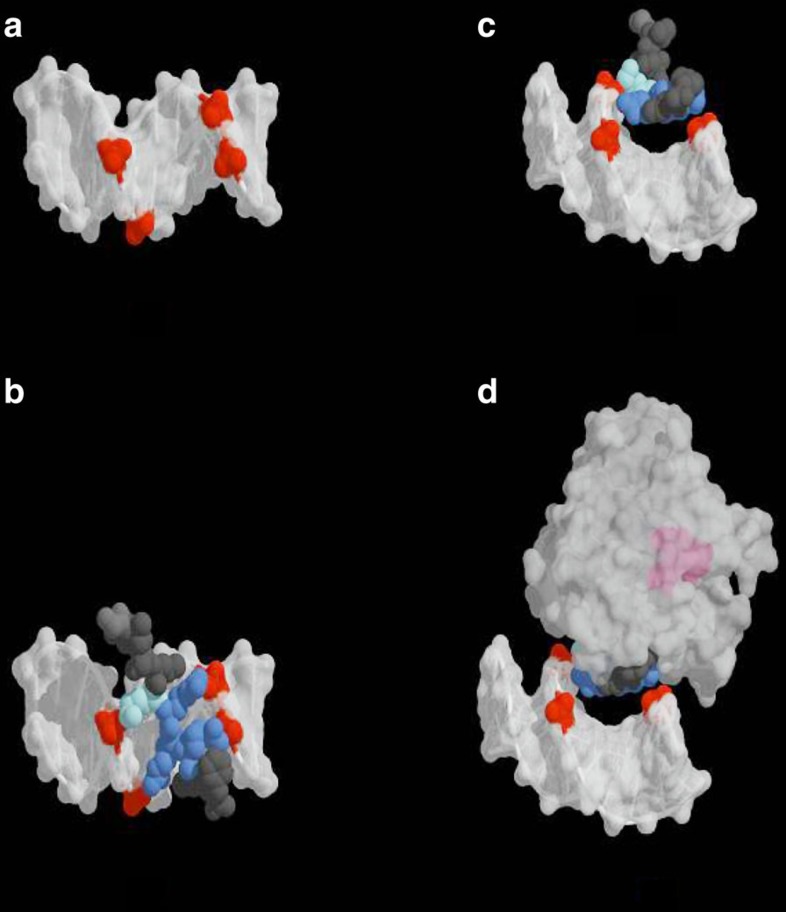
Figure 3. Model for the binding of pVIc and AVP–pVIc complexes to DNA.
(a) The structure of a B-form DNA dodecamer (PDB ID: 1HQ7) is displayed with four of its phosphate groups coloured in red. (b) The structure of pVIc obtained from the crystal structure of the AVP–pVIc complex (PDB ID: 1NLN) is shown docked to the DNA. The four basic residues of pVIc, the one lysine residue and the three arginine residues, are coloured in light blue and dark blue, respectively. (c) The DNA-pVIc complex was rotated ∼90° on its x axis to show the contacts of the peptide with the major groove. (d) The AVP–pVIc structure is displayed, showing that DNA binding is dominated by the pVIc moiety. The majority of the binding enthalpy between the protein–peptide complex and DNA is likely to originate through electrostatic interactions between the four, contiguous, basic residues of pVIc (KRRR, in blue) in the AVP–pVIc complex and phosphate groups (in red) in the backbone of the dsDNA. The active site of AVP is coloured pink, showing that the proteinase active site is sterically unhindered by DNA binding, consistent with the proteinase being able to bind to and cleave substrates while bound to and sliding on DNA. The figure was rendered using PyMol (The PyMOL Molecular Graphics System, Version 1.2r.3pre, Schrödinger, LLC).