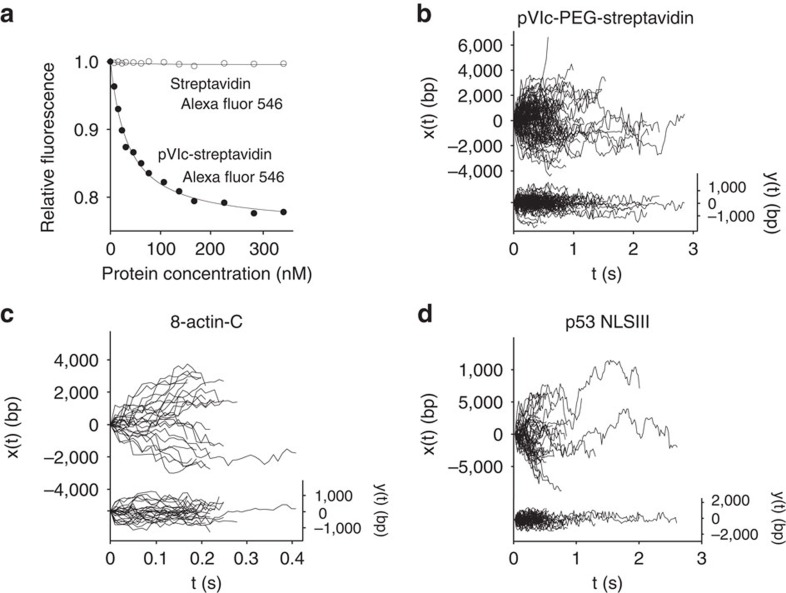
Figure 4. Sliding of heterologous proteins and peptides.
(a) The equilibrium dissociation constant for the binding of (pVIc-biotin)-streptavidin to 18-mer dsDNA was determined by fluorescence resonance energy transfer. The quenching of the fluorescence intensity of the donor molecule, fluorescein-labelled 18-bp dsDNA, as a function of the concentration of the acceptor molecule (pVIc-biotin)-streptavidin Alexa Fluor 546 is shown by the closed circles. The relative fluorescence intensity is the fluorescence intensity at a specific concentration of acceptor divided by the initial fluorescence intensity of the donor in the absence of acceptor. The line through the closed circles represents the nonlinear regression fit of the experimental data to a 1:1 ligand–receptor model. The open circles represent data from the titration of 10 nM fluorescein-labelled 18-mer dsDNA with streptavidin Alexa Fluor 546. These data indicated that streptavidin Alex Fluor 546 did not bind to DNA. (b) (pVIc-biotin)-streptavidin complexes diffuse rapidly along DNA (x(t), left axis, 106 trajectories). Motion transverse to the DNA (y(t), right axis, is represented on the same scale, as a control. (c) A peptide (SIVHRKCF) with the last 8 amino acids of β-actin diffuses rapidly (69 trajectories) along DNA x(t). (d) The 13-amino acid peptide of NLSIII of the p53 protein (STSRHKKLMFKTE) diffuses rapidly along DNA at pH 6.5 (45 trajectories).