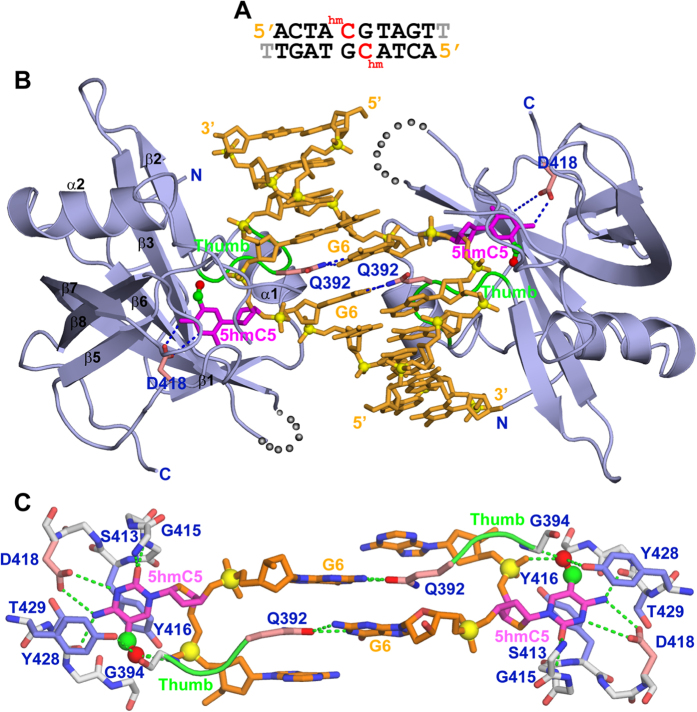
Figure 2. Crystal structure of the SUVH5 SRA domain bound to the fully-5 hmCG DNA.
(A) Sequence of the 10-mer self-complementary fully-5 hmCG DNA (with a 3′-T overhang) containing 5 hmC-G base pairs in the centre of the duplex. (B) Stick (DNA) and ribbon (protein) representation of the 2.6 Å crystal structure of the 2:1 SUVH5 SRA-5 hmCG DNA duplex complex. The DNA is shown in orange, except for 5 hmC5, which is shown in magenta. The methylene and hydroxyl groups are shown as small green and red spheres, respectively, in Figs 2, 3 and 5. The backbone phosphorus atoms are shown as yellow balls, and the 5′ and 3′ ends of the DNA are labelled. The SRA domain is shown in blue, with its secondary structural elements labelled with the same α/β numbering scheme as that of the UHRF1 SRA10. The thumb loop is in green. The 5 hmC5s residues on the partner strands are flipped out of the minor groove and are positioned in the binding pockets of the individual SRA domains. The Watson-Crick edge of 5 hmC5 is hydrogen bonded with the side chain of Asp418. The side chain of Gln392 inserts into and fills the gap created by the flipped-out 5 hmC base and pairs with the Watson-Crick edge of the G6 base. (C) Magnified view of the relative alignments of the G6-Gln392 interactions and the interaction of the flipped-out symmetrical 5 hmC5s with the residues lining the binding pocket. Gln392 from the thumb loop is inserted into and fills the gap created by the flipped-out 5 hmC base, and its side chain forms stacking interactions with the flanking bases. The 5 hmC5 base is positioned between the aromatic rings of Tyr416 and Tyr428; its Watson-Crick edge is hydrogen bonded to the protein backbone and side chain of Asp418. The hydroxyl group at fifth position of 5 hmC forms intermolecular interactions with the side chain of Tyr428 and an amide group of G394 from the thumb and intramolecular interactions with the phosphate group.